第 10 章 LI-6800 的数据分析
10.1 数据格式
此处参考 @ref(batch_question) 相关内容即可。
10.2 LI-6800 与 LI-6400 使用时的差别
plantecophys 使用时建立在 LI-6400XT 基础之上的软件包,因此在 LI-6800 代码中,需要改动的是 fitaci、fitacis 及 fitBB 中的 varnames 选项,也就是将 LI-6400XT 的表头改为 LI-6800 的表头。
以 fitaci 函数为例:
fitaci(aci, varnames =
list(ALEAF = "A", Tleaf = "Tleaf", Ci = "Ci",
PPFD = "Qin", Rd = "Rd"))注:我个人觉得这样使用对于 LI-6800 来讲十分不方便,修改了一下
plantecophys,专门给 LI-6800 使用,具体可参考:
1.使用示例
10.3 光响应曲线注意事项
光响应曲线的拟合相对简单,仅需要光强和光合速率的值,其中需要修改的部分仅为光强的赋值部分,在文件名一致的前提下,修改如下代码即可:
lrc_Q <- lrc$Qin
lrc_A <- lrc$A 10.5 LI-6800 RACiR的测量与拟合
在评估作物性状时,V\(_{cmax}\) 及 J\(_{max}\)时非常有用,传统的 A–Ci 曲线测量要求植物叶片要在一定浓度 CO\(_{2}\) 下适应几分钟后完成测量,这样的测量有几个缺点:
- 测量时间长,一条曲线至少需要 20 – 30 min,样本量多,重复多时,这种方法几乎没有可行性。
- 整个测量过程中,时间长,酶的激活状态会有变化,叶绿体会移动,气孔的开度也会发生变化。
而 LI-6800 独有的 auto control 功能在算法上允许用户自定义 CO\(_{2}\) 的起始浓度和种植浓度、变化方式(线性或其他)、所花费的时间,再加上其 IRGAs 极快的响应频率,使得短时间内的 A–Ci 的测量成为现实,即快速 CO\(_{2}\) 响应曲线 RACiR 测量实验,该功能使得 5 min 内测量 A–Ci 曲线成为可能。该方法的实现可参考 J. R. Stinziano et al. (2017) 的文章。
Joseph R. Stinziano et al. (2018) 针对 RACiR技术的疑问做了解答并提出了准确测量的建议,概括如下:
- 首先,采用 100 ppm/min 的变化速率是与标准方法重合度最高的测量。
- 其次,明确研究问题,目前已有研究表明Vcmax 与 Jmax 的计算结果与标准测量方法结果无显著差异。
- 任何条件的改变,都需要做空叶室校准,例如:流速,气体浓度变化方向、温度,斜率等。
- 空叶室校准与叶片测量采用严格的同一次校准,因为 IRGA 的漂移,需要再次匹配时,或者环境条件改变时,需要重新做空叶室校准。是否需要匹配,可通过不加叶片的最初状态查看,此时 A 值应接近为0,reference 和 sample 气体浓度读数接近相等。
- IRGA 分析器使用 5 此多项式进行校准,推荐使用 1 次到 5 次多项式进行拟合,然后根据 BIC 指数来确定最合适的空叶室校准系数(即非参数拟合的模型选择的问题)。 确定最合适的浓度变化范围。通常需要去掉最初和最后 30 s的数据。
- 最小化校准和测量值之间的水分摩尔分数差异。甚至有可能需要控制 reference 或 sample 的水的摩尔分数而不是 Vpdleaf。 通过预实验来确定最合适的 \(CO_2\) 变化范围和随时间的斜率。
本文之后关于racir 的空叶室校正部分全部不再支持,因为LI-6800 1.5之后的系统版本,关于两个分析器在快速变化环境条件下的校正均由仪器完成,分析工作直接使用 plantecophys 等软件拟合即可。
例如下面的数据采用了 1.5.02 的 BP 程序 DAT_CO2_Continuous.py,直接测量的 RACiR 数据,结果如下:
remotes::install_github("zhujiedong/plantecophys2")adyn <- read.csv("data/racir_adyn.csv")
adyn_fit <- plantecophys2::fitaci(adyn)## Registered S3 methods overwritten by 'plantecophys2':
## method from
## coef.BBfit plantecophys
## coef.BBfits plantecophys
## coef.acifit plantecophys
## coef.acifits plantecophys
## fitted.acifit plantecophys
## plot.acifit plantecophys
## plot.acifits plantecophys
## print.BBfit plantecophys
## print.BBfits plantecophys
## print.acifit plantecophys
## print.acifits plantecophys
## summary.acifit plantecophys##
## Formula: ALEAF ~ acifun_wrap(Ci, PPFD = PPFD, Vcmax = Vcmax, Jmax = Jmax,
## Rd = Rd, Tleaf = Tleaf, Patm = Patm, TcorrectVJ = Tcorrect,
## alpha = alpha, theta = theta, gmeso = gmeso, EaV = EaV, EdVC = EdVC,
## delsC = delsC, EaJ = EaJ, EdVJ = EdVJ, delsJ = delsJ, Km = Km,
## GammaStar = GammaStar)
##
## Parameters:
## Estimate Std. Error t value Pr(>|t|)
## Vcmax 26.99477 0.11547 233.79 <2e-16 ***
## Jmax 46.99725 0.11780 398.96 <2e-16 ***
## Rd 1.38036 0.02146 64.31 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.1347 on 295 degrees of freedom
##
## Number of iterations to convergence: 7
## Achieved convergence tolerance: 5.627e-06plot(adyn_fit)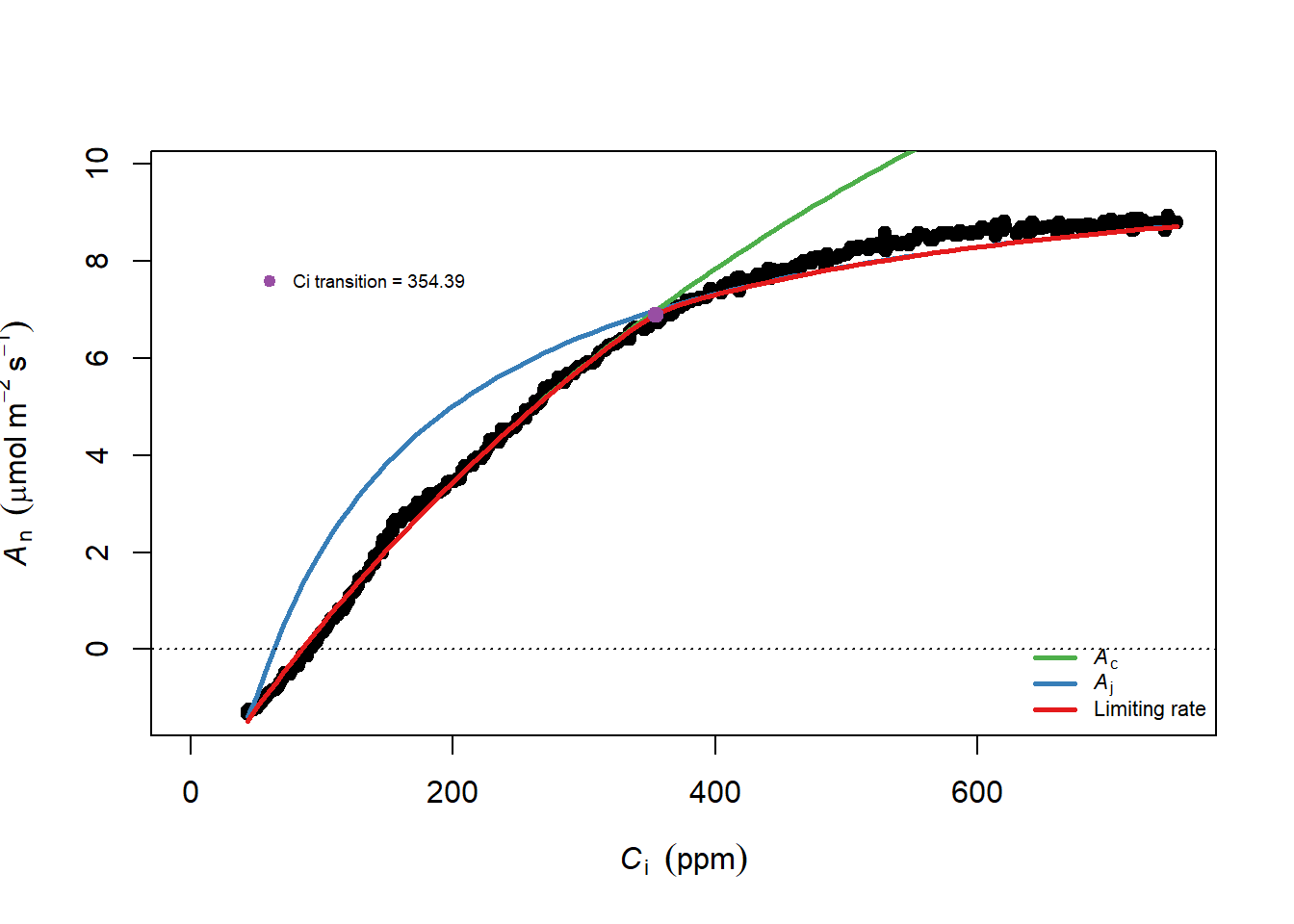
(#fig:adyn_plot)使用 Adyn 来进行RACiR 的直接测量数据
@ref(fig: dyn_plot) 展示的使用 LI-6800 自带程序测量的数据,无需修正,测量作物为早春温室的水稻。
** 以上为推荐方式,之前的内容自本日(2021.11.12)起正式删除 **
10.6 LI-6800 RACiR簇状叶的测量与拟合
该部分内容请使用 DAT 来测量,实际上后来的
DAT_CO2_Continuous.py的测量方式参考了这篇文献的内容。代码部分删除,因为新方法无需要再进行校准了。
Coursolle et al. (2019) 测量了簇状叶黑云杉和香脂冷杉两种簇状叶植物的 RACiR,其中的试验方法和结论值得在测量时借鉴,测量方法上:
簇状叶室体积远大于荧光叶室和其他叶室,使用的 \(CO_2\) 的变化为: 15 min 内从 20 ppm 到 1520 ppm 的变化,即变化的速率为 \(100 ppm \cdot min^{-1}\)。但也测试了 200 - 800 ppm的部分曲线。
拟合使用了测量的 Rd,测量方法为:控制 reference 气路在 420 ppm 的 \(CO_2\) 和 22 \(mmol \cdot mol^{-1}\) 的 H2O 浓度,控制温度为 25 C,诱导后测量 Rd。
得到了一些有帮助的结论:
使用更大的叶室测量 RACiR 是可行的(36 \(cm^2\)),叶室环境的控制需要通过预实验来确定。
该实验使用的 ACi 曲线测量时间在 30 到 36 min,而 RACiR 使用的完整的二氧化碳的浓度范围时,曲线耗时最大的时间接近 22 min。但使用 200 - 800 ppm 范围的变化,则时间可以下降 50%,这些部分范围的测量则可以应用于植物胁迫和表型平台的研究。
实验结果证明只要 match 的调整值保持不变即无需进行空叶室校准(也就是无需匹配的意思,实际的时间间隔取决于仪器的状态),但最新的 range match 功能可有效的增加空叶室校准的时间间隔(新功能,作者试验时尚未推出该功能)。
作者建议最好测量暗呼吸的速率,以获得最佳的 Vcmax 和 Jmax 计算结果。如果有第二台光合仪来测量则可有效的缩短测量时间。
10.7 多个速率的 RACiR 曲线研究
自 RACiR 技术诞生以来,极大的缩短了 Vcmax 以及 Jmax 的测量时间(J. R. Stinziano et al. (2017)),但也引起了一系列争议,作者也对业内的质疑进行了一一的解答 (Joseph R. Stinziano et al. (2018)),但除了因为时间长短导致酶活性,叶绿体位置等差异外,RACiR 还能说明哪些问题呢?Joseph R., Rachael K., and David T. (2019) 最新的研究给出一系列结论:
扩散限制(\(CO_2\) 总导度) 和光呼吸导致了表观上的标准 ACi 曲线和 RACiR 测量之间的偏差,表明他们的差异是由生物因子引起,而非仪器导致的人为误差。
上述原因导致的二者之间的偏差,如果不进行修正,那么将显著的低估 \(\Gamma^*\), 除非使用多个速率的 RACiR 来修正。
较高速率的 RACiR 曲线会增大其与标准曲线之间的偏差,但这个差距在无光呼吸的条件下会减小。
因为光呼吸和气体扩散限制与物种相关,结合以上结论,可以使用多个速率的 RACiR 来估算对 \(CO_2\) 的总导度以及相对量的光呼吸速率。
一些可能的方向:
扩散限制影响 Cc 速率的变化,说明对具有较高总阻力与 \(CO_2\) 比值的物种,例如针叶物种,C4 植物,较高的阻力导致 RACiR 与 标准 ACi 测量斜率更大的差异,或者测量的前提假设被破坏。
RACiR 可检测到代谢中 \(CO_2\) 的滞后性,各种滞后性的检测对标准 ACi 测量也具有指示性。
文中利用 R 实现了光呼吸之后模型和气体扩散限制模型,本文内容主要对文献中附录材料的源码进行解释:
10.7.1 光呼吸滞后模型
为测试光呼吸的滞后性,作者使用一系列预先设定的参数,模拟了一条 ACc曲线,假定 Rubisco 激活状态为 100%,并且在整个测量过程中气孔导度是不变的。然后使用这些参数来模拟 RACiRs 曲线,并且假定光呼吸分别需要 0, 15, 30, 60, 120 或 300 s 来对变化的 \(CO_2\) 进行响应,在实际效果上,这意味着 Cc 在最初的 0, 15, 30, 60, 120 或 300 内是不变的,最后我们对 \(\Gamma^*\) 和 Ci 使用线性回归进行计算。
10.7.1.1 构造基础数据
模型第一步,则是对需要使用的参数,根据文献和实际情况进行赋值,具体内容参考代码注释。
library(ggplot2)
library(plyr)##
## Attaching package: 'plyr'## The following object is masked from 'package:purrr':
##
## compactlibrary(gridExtra)
# 对图例使用自定义颜色
gg_color_hue <- function(n) {
hues = seq(15, 375, length = n + 1)
hcl(h = hues, l = 65, c = 100)[1:n]
}
#Maximum Rubisco carboxylation rate in umol m-2 s-1
Vcmax <- 110
#Maximum Rubisco oxygenation rate in umol m-2 s-1;
#Ratio from Bernacchi et al. 2001. PCE 24:253-259
Vomax <- 0.29 * Vcmax
#Dark respiration in umol m-2 s-1
R <- 2
#Michaelis-Menten constant for Rubisco carboxylation in umol mol-1
Kc <- 404.9
#Michaelis-Menten constant for Rubisco oxygenation in mmol mol-1
Ko <- 278.4
#oxygen concentration in mmol mol-1
O2 <- 210
Kco <- Kc * (1 + O2 / Ko)
#Boundary layer conductance in mol m-2 s-1
BLC <- 2
#stomatal conductance in mol m-2 s-1
gsw <- 0.4
#mesophyll conductance in mol m-2 s-1
gm <- 1
#Chloroplastic CO2 in umol mol-1
Cc <- as.numeric(c(25:400))
#oxygenation rate in umol m-2 s-1
vo <- Vomax * O2 / (O2 + Ko * (1 + Cc / Kc))
#carboxylation rate in umol m-2 s-1
vc <- Vcmax * (Cc) / (Cc + Kco)
#Net CO2 assimilation in umol m-2 s-1
A <- vc - 0.5 * vo - R
#Apparent CO2 assimilation rate in umol m-2 s-1
Aapparent <- vc - 0.5 * vo
#Intercellular CO2 in umol mol-1
Ci <- A / gm + Cc
#Boundary layer CO2 in umol mol-1
Cb <- A / gsw + Ci
#Reference CO2 in umol mol-1
Cr <- A / BLC + Cb
#根据Cc浓度的个数,构造向量
Counter <- as.numeric(c(1:length(Cc)))
#也就是以秒计算的cr与时间的模型
RateCrmodel <- lm(Cr ~ Counter)
#转化为分钟的cr的斜率
RateCr <- coef(RateCrmodel)[2] * 60
#转换为分钟的边界层导度斜率
RateCbmodel <- lm(Cb ~ Counter)
RateCb <- coef(RateCbmodel)[2] * 60
#转换为分钟的ci的斜率
RateCimodel <- lm(Ci ~ Counter)
RateCi <- coef(RateCimodel)[2] * 60
#转换为分钟的Cc的斜率
RateCcmodel <- lm(Cc ~ Counter)
RateCc <- coef(RateCcmodel)[2] * 60 10.7.2 光呼吸滞后性代码
下面代码的目的是为得到 ACi 响应曲线受光呼吸延迟的影响,尤其是在临近补偿点时。
延迟模块
#假定有15s延迟时的数据,即相比上面构造的Cc数据减少15个点
Cc15 <- as.numeric(c((min(Cc) + 15):max(Cc), rep(max(Cc), 15)))
vo15 <- Vomax * O2 / (O2 + Ko * (1 + Cc15 / Kc))
A15 <- vc - 0.5 * vo15 - R
Aapparent15 <- vc - 0.5 * vo15
#30 s 延迟数据
Cc30 <- as.numeric(c((min(Cc) + 30):max(Cc), rep(max(Cc), 30)))
vo30 <- Vomax * O2 / (O2 + Ko * (1 + Cc30 / Kc))
A30 <- vc - 0.5 * vo30 - R
Aapparent30 <- vc - 0.5 * vo30
#60s 延迟数据
Cc60 <- as.numeric(c((min(Cc) + 60):max(Cc), rep(max(Cc), 60)))
vo60 <- Vomax * O2 / (O2 + Ko * (1 + Cc60 / Kc))
A60 <- vc - 0.5 * vo60 - R
Aapparent60 <- vc - 0.5 * vo60
#120s延迟数据
Cc120 <- as.numeric(c((min(Cc) + 120):max(Cc), rep(max(Cc), 120)))
vo120 <- Vomax * O2 / (O2 + Ko * (1 + Cc120 / Kc))
A120 <- vc - 0.5 * vo120 - R
Aapparent120 <- vc - 0.5 * vo120
#300s延迟数据
Cc300 <- as.numeric(c((min(Cc) + 300):max(Cc), rep(max(Cc), 300)))
vo300 <- Vomax * O2 / (O2 + Ko * (1 + Cc300 / Kc))
A300 <- vc - 0.5 * vo120 - R
Aapparent300 <- vc - 0.5 * vo300 10.7.3 数据的构造
下面的代码主要是将上文最终计算的数据构造数据集,并导出。
Anet <- c(A, A15, A30, A60, A120, A300)
Aapp <-
c(Aapparent,
Aapparent15,
Aapparent30,
Aapparent60,
Aapparent120,
Aapparent300)
Ccfull <- rep(Cc, 6)
Cifull <- rep(Ci, 6)
Delay <-
c(
rep("0", length(A)),
rep("15", length(A15)),
rep("30", length(A30)),
rep("60", length(A60)),
rep("120", length(A120)),
rep("300", length(A300))
)
PRdata <- as.data.frame(cbind(Anet, Aapp, Ccfull, Cifull, Delay))
write.csv(PRdata, "./data/PRdata.csv")10.7.4 光呼吸滞后性作图
下面的代码是将光呼吸的数据进行作图。
data <- read.csv("./data/PRdata.csv")
data$Ccfull <- as.numeric(data$Ccfull)
data$Delay <- as.factor(data$Delay)
# 净光合速率与Cc作图
AnetCc <- ggplot(data, aes(x = Ccfull, y = Anet, colour = Delay)) +
geom_point() +
labs(x = expression(C[c] ~ "(" * mu * mol ~ mol ^ {
-1
} * ")"),
y = expression(A[net] ~ "(" * mu * mol ~ m ^ {
-2
} ~ s ^ {
-1
} * ")")) +
labs(colour = 'Delay (s)') +
scale_x_continuous(limits = c(25, 100),
breaks = c(25, 40, 55, 70, 85, 100)) +
scale_y_continuous(limits = c(-5, 5)) +
scale_colour_brewer(palette = 'Spectral') +
#补偿点的参考线
geom_hline(yintercept = 0) +
theme_bw() +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank())
AnetCc## Warning: Removed 1848 rows containing missing values (geom_point).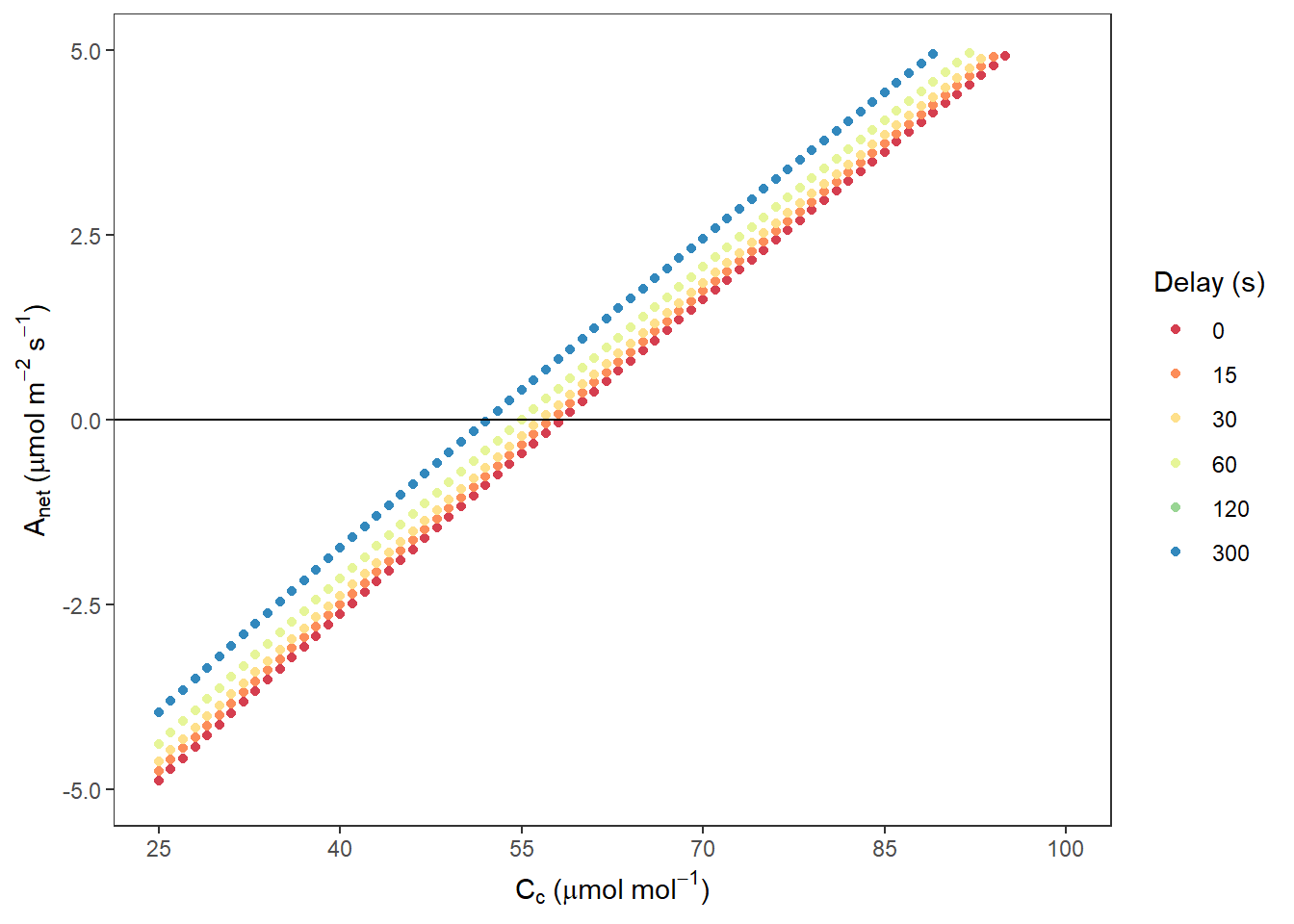
图 10.1: Anet VS. Cc
#净光合速率与Ci作图
AnetCi <- ggplot(data, aes(x = Cifull, y = Anet, colour = Delay)) +
geom_point() +
labs(x = expression(C[i] ~ "(" * mu * mol ~ mol ^ {
-1
} * ")"),
y = expression(A[net] ~ "(" * mu * mol ~ m ^ {
-2
} ~ s ^ {
-1
} * ")")) +
labs(colour = 'Delay (s)') +
scale_x_continuous(limits = c(25, 100),
breaks = c(25, 40, 55, 70, 85, 100)) +
scale_y_continuous(limits = c(-5, 5)) +
scale_colour_brewer(palette = 'Spectral') +
geom_hline(yintercept = 0) +
theme_bw() +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank())
AnetCi## Warning: Removed 1878 rows containing missing values (geom_point).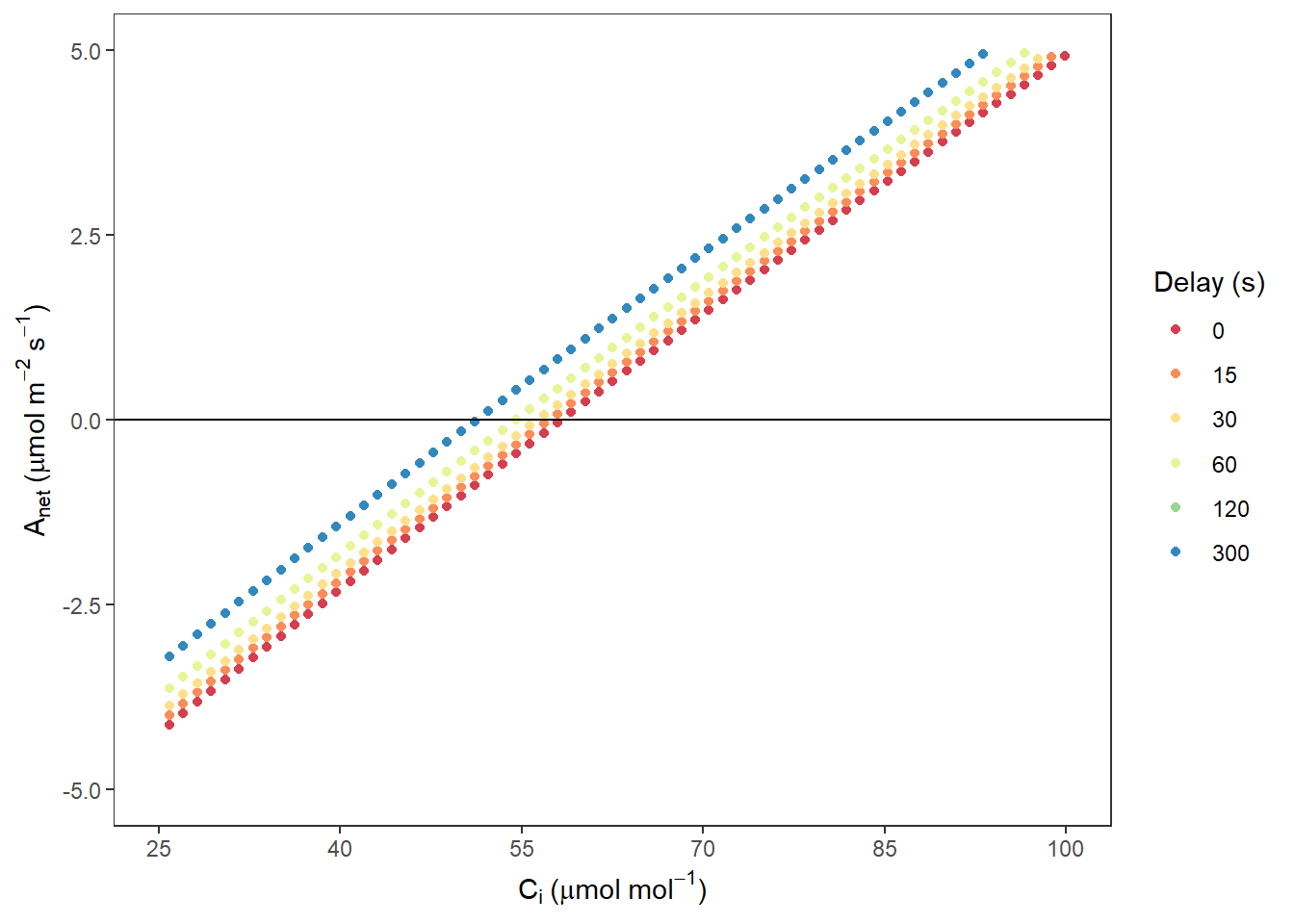
图 10.2: Anet VS. Ci
#表观光合与Cc作图
AappCc <- ggplot(data, aes(x = Ccfull, y = Aapp, colour = Delay)) +
geom_point() +
labs(x = expression(C[c] ~ "(" * mu * mol ~ mol ^ {
-1
} * ")"),
y = expression(A[apparent] ~ "(" * mu * mol ~ m ^ {
-2
} ~ s ^ {
-1
} * ")")) +
labs(colour = 'Delay (s)') +
scale_x_continuous(limits = c(25, 75),
breaks = c(25, 35, 45, 55, 65, 75)) +
scale_y_continuous(limits = c(-5, 5)) +
scale_colour_brewer(palette = 'Spectral') +
geom_hline(yintercept = 0) +
theme_bw() +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank())
AappCc## Warning: Removed 1959 rows containing missing values (geom_point).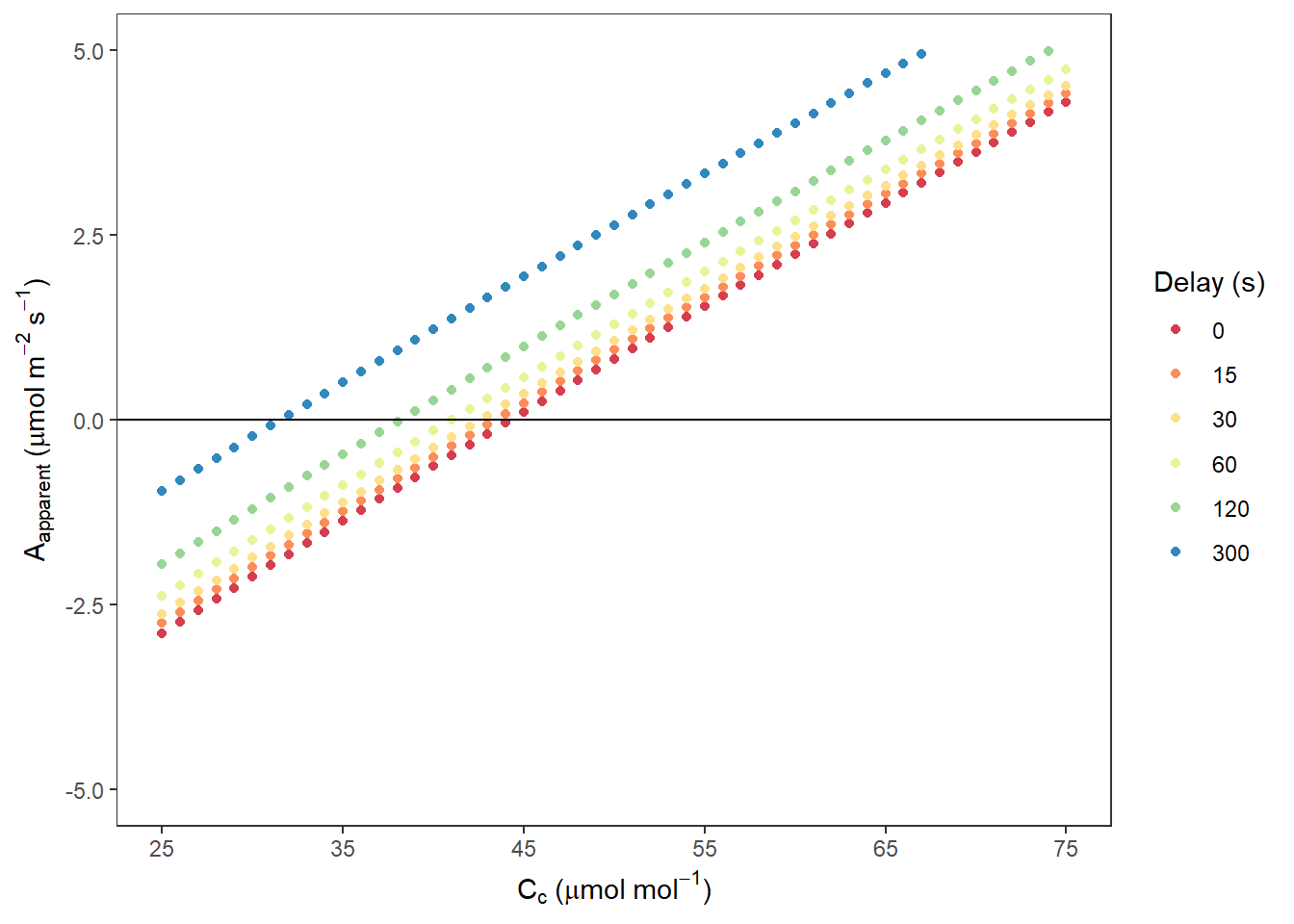
图 10.3: Aapparent VS. Cc
#表观与Ci作图
AappCi <- ggplot(data, aes(x = Cifull, y = Aapp, colour = Delay)) +
geom_point() +
labs(x = expression(C[i] ~ "(" * mu * mol ~ mol ^ {
-1
} * ")"),
y = expression(A[apparent] ~ "(" * mu * mol ~ m ^ {
-2
} ~ s ^ {
-1
} * ")")) +
labs(colour = 'Delay (s)') +
scale_x_continuous(limits = c(25, 75),
breaks = c(25, 35, 45, 55, 65, 75)) +
scale_y_continuous(limits = c(-5, 5)) +
scale_colour_brewer(palette = 'Spectral') +
geom_hline(yintercept = 0) +
theme_bw() +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank())
AappCi## Warning: Removed 2003 rows containing missing values (geom_point).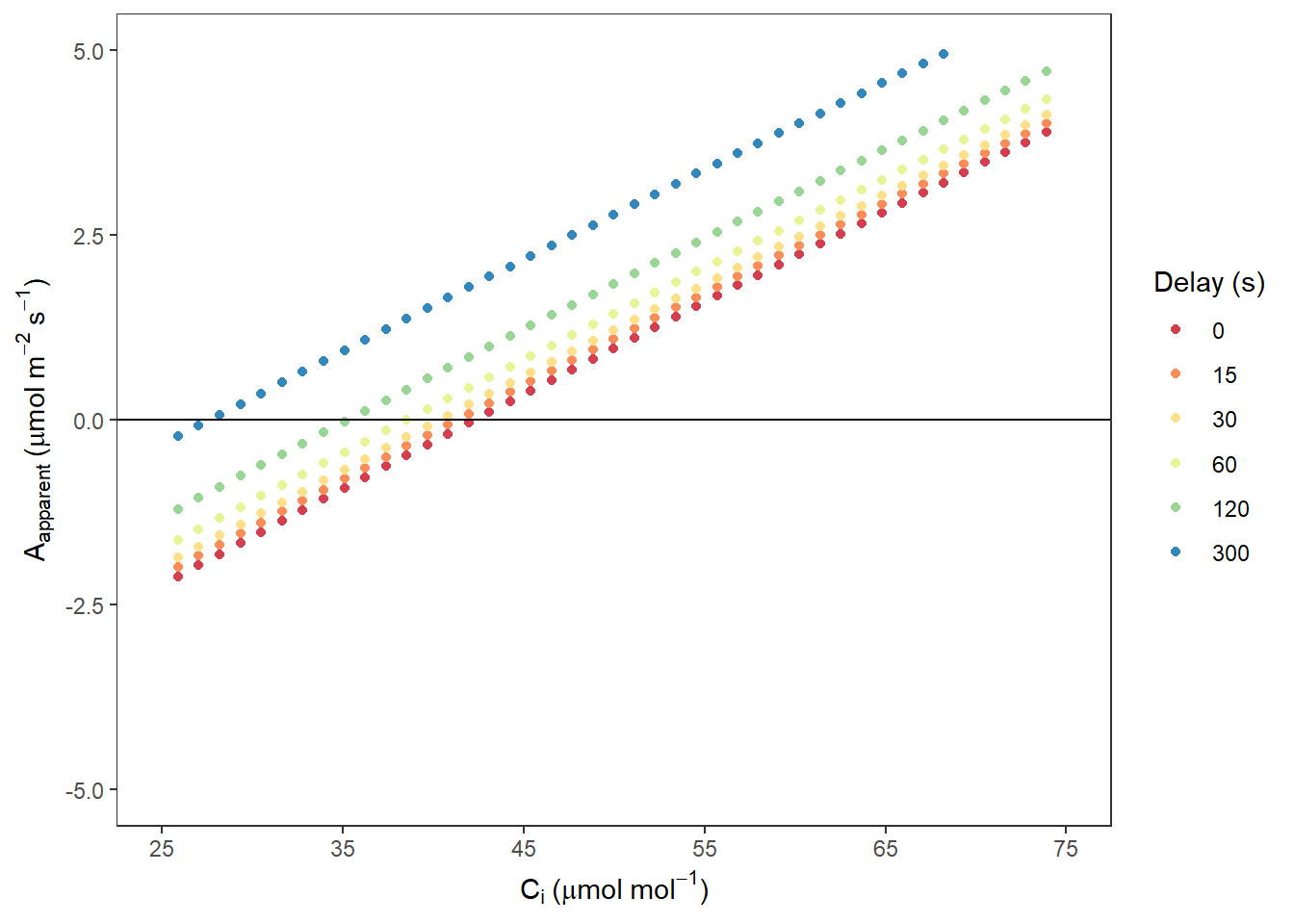
图 10.4: Aapparent VS. Ci
10.7.5 补偿点计算
计算不同的光呼吸时间延迟下的补偿点(基于Ci):
#对于基于Ci的数据,仅采用ci<100时的数据
dataCi <- data[data$Cifull < 100,]
dataCi0 <- dataCi[dataCi$Delay == "0",]
dataCi15 <- dataCi[dataCi$Delay == "15",]
dataCi30 <- dataCi[dataCi$Delay == "30",]
dataCi60 <- dataCi[dataCi$Delay == "60",]
dataCi120 <- dataCi[dataCi$Delay == "120",]
dataCi300 <- dataCi[dataCi$Delay == "300",]
#光呼吸无延迟时的计算,线性拟合
m1 <- lm(dataCi0$Anet ~ dataCi0$Cifull)
summary(m1)##
## Call:
## lm(formula = dataCi0$Anet ~ dataCi0$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.13165 -0.04518 0.01711 0.05408 0.06699
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -7.2248105 0.0199353 -362.4 <2e-16 ***
## dataCi0$Cifull 0.1228632 0.0003089 397.8 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.06081 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.582e+05 on 1 and 69 DF, p-value: < 2.2e-16#补偿点为截距比斜率(纵坐标为零)
Gamma0 <- -m1$coefficients[1] / m1$coefficients[2]
#光呼吸延时15s
m2 <- lm(dataCi15$Anet ~ dataCi15$Cifull)
summary(m2)##
## Call:
## lm(formula = dataCi15$Anet ~ dataCi15$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.13119 -0.04502 0.01705 0.05389 0.06676
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -7.0873326 0.0198665 -356.7 <2e-16 ***
## dataCi15$Cifull 0.1225900 0.0003078 398.3 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.0606 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.586e+05 on 1 and 69 DF, p-value: < 2.2e-16Gamma15 <- -m2$coefficients[1] / m2$coefficients[2]
#光呼吸延时30s
m3 <- lm(dataCi30$Anet ~ dataCi30$Cifull)
summary(m3)##
## Call:
## lm(formula = dataCi30$Anet ~ dataCi30$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.13076 -0.04488 0.01699 0.05372 0.06654
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -6.9553180 0.0198028 -351.2 <2e-16 ***
## dataCi30$Cifull 0.1223321 0.0003068 398.7 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.0604 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.59e+05 on 1 and 69 DF, p-value: < 2.2e-16Gamma30 <- -m3$coefficients[1] / m3$coefficients[2]
#光呼吸延时60s
m4 <- lm(dataCi60$Anet ~ dataCi60$Cifull)
summary(m4)##
## Call:
## lm(formula = dataCi60$Anet ~ dataCi60$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.13001 -0.04462 0.01690 0.05341 0.06616
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -6.706429 0.019689 -340.6 <2e-16 ***
## dataCi60$Cifull 0.121858 0.000305 399.5 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.06006 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.596e+05 on 1 and 69 DF, p-value: < 2.2e-16Gamma60 <- -m4$coefficients[1] / m4$coefficients[2]
#光呼吸延时120s
m5 <- lm(dataCi120$Anet ~ dataCi120$Cifull)
summary(m5)##
## Call:
## lm(formula = dataCi120$Anet ~ dataCi120$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.12879 -0.04421 0.01673 0.05292 0.06555
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -6.2616980 0.0195062 -321.0 <2e-16 ***
## dataCi120$Cifull 0.1210486 0.0003022 400.5 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.0595 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.604e+05 on 1 and 69 DF, p-value: < 2.2e-16Gamma120 <- -m5$coefficients[1] / m5$coefficients[2]
#光呼吸延时300s
m6 <- lm(dataCi300$Anet ~ dataCi300$Cifull)
summary(m6)##
## Call:
## lm(formula = dataCi300$Anet ~ dataCi300$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.12879 -0.04421 0.01673 0.05292 0.06555
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -6.2616980 0.0195062 -321.0 <2e-16 ***
## dataCi300$Cifull 0.1210486 0.0003022 400.5 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.0595 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.604e+05 on 1 and 69 DF, p-value: < 2.2e-16Gamma300 <- -m6$coefficients[1] / m6$coefficients[2]构造数据并作图
GammaCi <- c(Gamma0, Gamma15, Gamma30, Gamma60, Gamma120, Gamma300)
ints <-
c(
m1$coefficients[1],
m2$coefficients[1],
m3$coefficients[1],
m4$coefficients[1],
m5$coefficients[1],
m6$coefficients[1]
)
slps <-
c(
0,
m2$coefficients[2] - m1$coefficients[2],
m3$coefficients[2] - m1$coefficients[2],
m4$coefficients[2] - m1$coefficients[2],
m5$coefficients[2] - m1$coefficients[2],
m6$coefficients[2] - m1$coefficients[2]
)
dels <- c(0, 15, 30, 60, 120, 300)
summary(lm(ints ~ dels))##
## Call:
## lm(formula = ints ~ dels)
##
## Residuals:
## (Intercept) (Intercept).1 (Intercept).2 (Intercept).3 (Intercept).4
## -0.20351 -0.11262 -0.02719 0.12853 0.38691
## (Intercept).5
## -0.17212
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -7.0213001 0.1343090 -52.277 8.01e-07 ***
## dels 0.0031057 0.0009959 3.119 0.0356 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.2503 on 4 degrees of freedom
## Multiple R-squared: 0.7086, Adjusted R-squared: 0.6357
## F-statistic: 9.725 on 1 and 4 DF, p-value: 0.03558plot(ints ~ dels)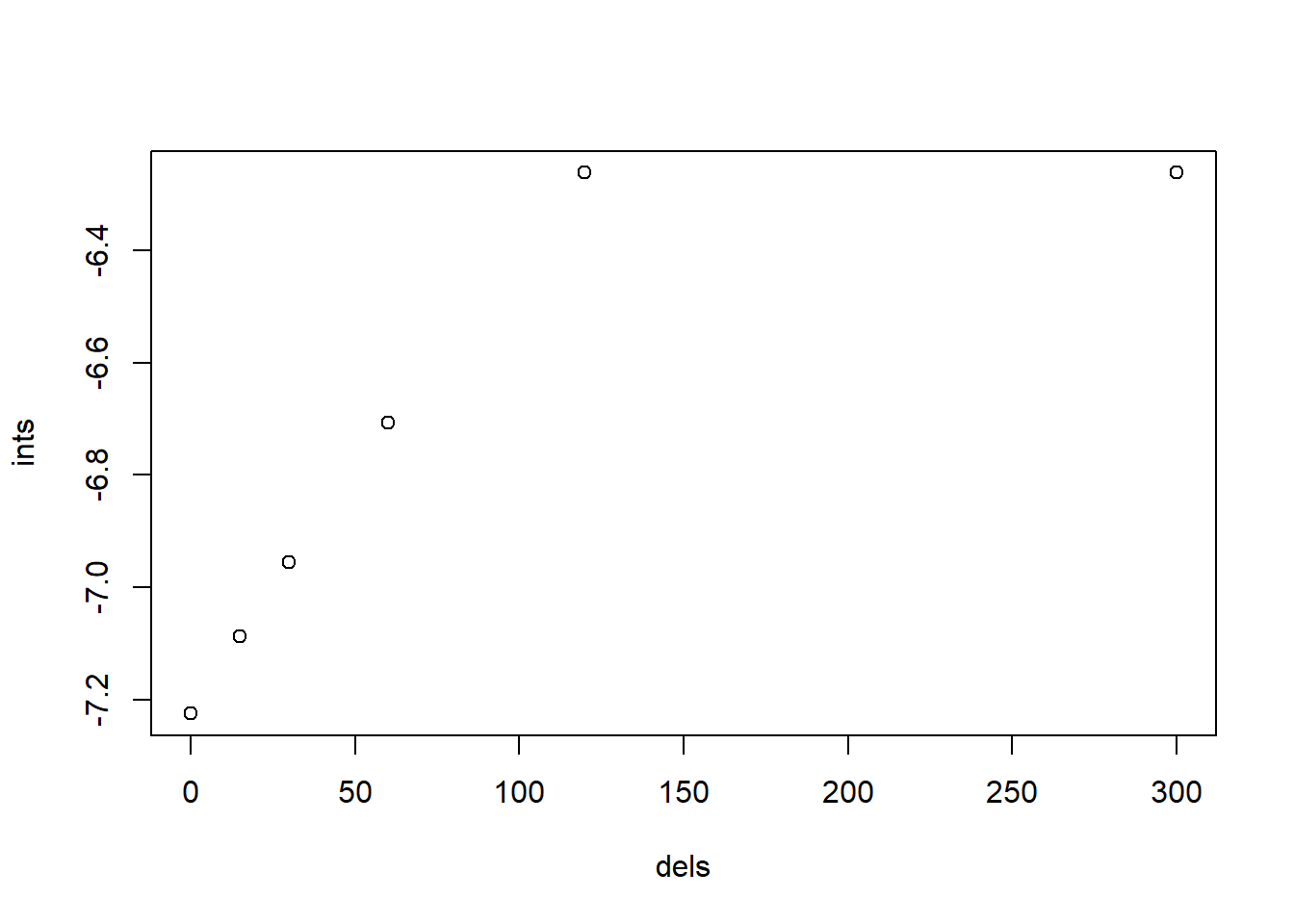
图 10.5: 基于 Ci 的不同延时下的截距
plot(slps ~ dels)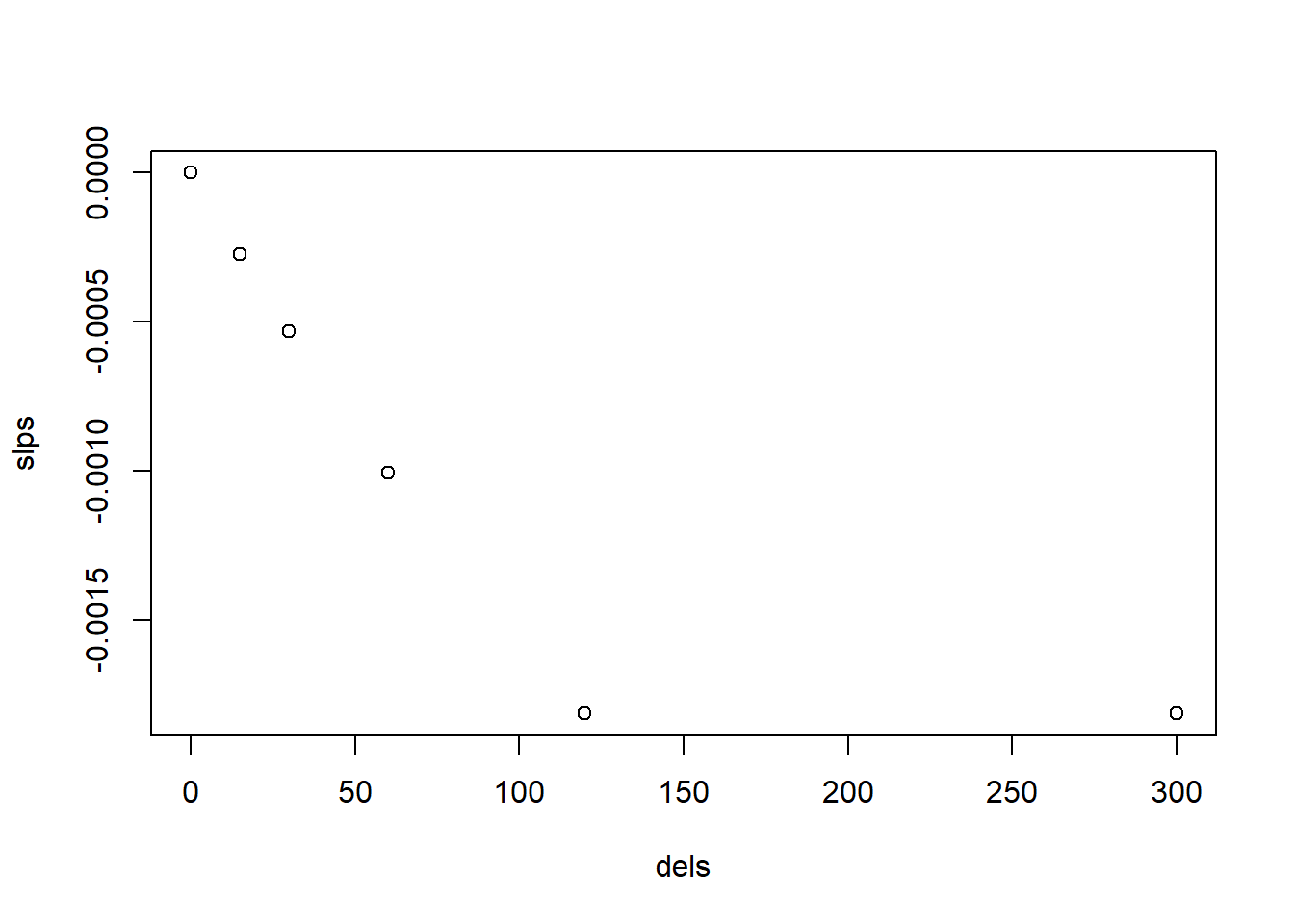
图 10.6: 基于 Ci 的不同延时下的斜率变化
summary(lm(slps ~ dels - 1))##
## Call:
## lm(formula = slps ~ dels - 1)
##
## Residuals:
## dataCi15$Cifull dataCi30$Cifull dataCi60$Cifull
## -2.711e-19 -1.574e-04 -2.995e-04 -5.424e-04
## dataCi120$Cifull dataCi300$Cifull
## -8.881e-04 5.015e-04
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## dels -7.72e-06 1.63e-06 -4.738 0.00516 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.0005383 on 5 degrees of freedom
## Multiple R-squared: 0.8178, Adjusted R-squared: 0.7814
## F-statistic: 22.44 on 1 and 5 DF, p-value: 0.005161基于 Cc 的补偿点计算结果:
# 仅使用 Cc < 75的数据点拟合,过程同ci
dataCc <- data[data$Ccfull < 75, ]
dataCc0 <- dataCc[dataCc$Delay == "0", ]
dataCc15 <- dataCc[dataCc$Delay == "15", ]
dataCc30 <- dataCc[dataCc$Delay == "30", ]
dataCc60 <- dataCc[dataCc$Delay == "60", ]
dataCc120 <- dataCc[dataCc$Delay == "120", ]
dataCc300 <- dataCc[dataCc$Delay == "300", ]
# 无延迟数据
m1 <- lm(dataCc0$Anet ~ dataCc0$Ccfull)
summary(m1)##
## Call:
## lm(formula = dataCc0$Anet ~ dataCc0$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.075648 -0.025444 0.009857 0.031706 0.039431
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -8.4062011 0.0181852 -462.3 <2e-16 ***
## dataCc0$Ccfull 0.1438710 0.0003527 407.9 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03599 on 48 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 1.664e+05 on 1 and 48 DF, p-value: < 2.2e-16Gamma0 <- -m1$coefficients[1] / m1$coefficients[2]
#延时15s数据
m2 <- lm(dataCc15$Anet ~ dataCc15$Ccfull)
summary(m2)##
## Call:
## lm(formula = dataCc15$Anet ~ dataCc15$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.075393 -0.025359 0.009824 0.031599 0.039299
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -8.2659145 0.0181243 -456.1 <2e-16 ***
## dataCc15$Ccfull 0.1435470 0.0003515 408.4 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03587 on 48 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 1.668e+05 on 1 and 48 DF, p-value: < 2.2e-16Gamma15 <- -m2$coefficients[1] / m2$coefficients[2]
#延时30s数据
m3 <- lm(dataCc30$Anet ~ dataCc30$Ccfull)
summary(m3)##
## Call:
## lm(formula = dataCc30$Anet ~ dataCc30$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.075158 -0.025280 0.009794 0.031501 0.039177
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -8.1312578 0.0180681 -450.0 <2e-16 ***
## dataCc30$Ccfull 0.1432414 0.0003504 408.8 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03576 on 48 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 1.671e+05 on 1 and 48 DF, p-value: < 2.2e-16Gamma30 <- -m3$coefficients[1] / m3$coefficients[2]
#延时60s数据
m4 <- lm(dataCc60$Anet ~ dataCc60$Ccfull)
summary(m4)##
## Call:
## lm(formula = dataCc60$Anet ~ dataCc60$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.074737 -0.025139 0.009739 0.031325 0.038959
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -7.8775340 0.0179675 -438.4 <2e-16 ***
## dataCc60$Ccfull 0.1426797 0.0003485 409.4 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03556 on 48 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 1.676e+05 on 1 and 48 DF, p-value: < 2.2e-16Gamma60 <- -m4$coefficients[1] / m4$coefficients[2]
#延时120s数据
m5 <- lm(dataCc120$Anet ~ dataCc120$Ccfull)
summary(m5)##
## Call:
## lm(formula = dataCc120$Anet ~ dataCc120$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.074060 -0.024912 0.009651 0.031042 0.038607
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -7.4246412 0.0178052 -417.0 <2e-16 ***
## dataCc120$Ccfull 0.1417238 0.0003453 410.4 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03524 on 48 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 1.684e+05 on 1 and 48 DF, p-value: < 2.2e-16Gamma120 <- -m5$coefficients[1] / m5$coefficients[2]
#延时300s数据
m6 <- lm(dataCc300$Anet ~ dataCc300$Ccfull)
summary(m6)##
## Call:
## lm(formula = dataCc300$Anet ~ dataCc300$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.074060 -0.024912 0.009651 0.031042 0.038607
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -7.4246412 0.0178052 -417.0 <2e-16 ***
## dataCc300$Ccfull 0.1417238 0.0003453 410.4 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03524 on 48 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 1.684e+05 on 1 and 48 DF, p-value: < 2.2e-16Gamma300 <- -m6$coefficients[1] / m6$coefficients[2]GammaCc <- c(Gamma0, Gamma15, Gamma30, Gamma60, Gamma120, Gamma300)
ints <-
c(
m1$coefficients[1],
m2$coefficients[1],
m3$coefficients[1],
m4$coefficients[1],
m5$coefficients[1],
m6$coefficients[1]
)
dels <- c(0, 15, 30, 60, 120, 300)
plot(ints ~ dels)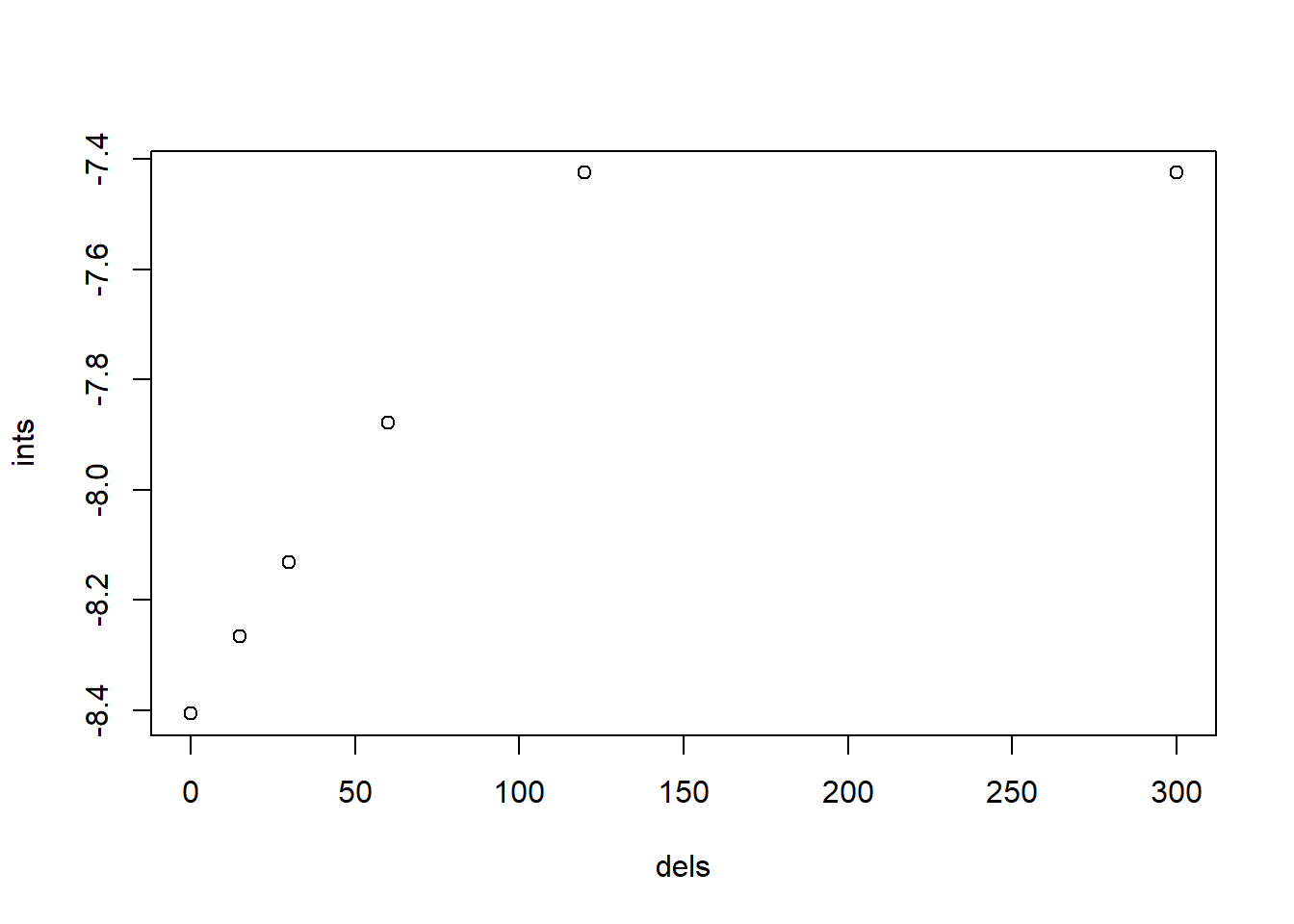
图 10.7: 基于 Cc 的不同延时下的时间
summary(lm(ints ~ dels))##
## Call:
## lm(formula = ints ~ dels)
##
## Residuals:
## (Intercept) (Intercept).1 (Intercept).2 (Intercept).3 (Intercept).4
## -0.20760 -0.11478 -0.02759 0.13119 0.39421
## (Intercept).5
## -0.17542
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -8.198602 0.136891 -59.891 4.65e-07 ***
## dels 0.003165 0.001015 3.118 0.0356 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.2552 on 4 degrees of freedom
## Multiple R-squared: 0.7085, Adjusted R-squared: 0.6356
## F-statistic: 9.72 on 1 and 4 DF, p-value: 0.03561#GammaStar
# For Ci-based estimates, only use Ci < 100
dataCi <- data[data$Cifull < 100,]
dataCi0 <- dataCi[dataCi$Delay == "0",]
dataCi15 <- dataCi[dataCi$Delay == "15",]
dataCi30 <- dataCi[dataCi$Delay == "30",]
dataCi60 <- dataCi[dataCi$Delay == "60",]
dataCi120 <- dataCi[dataCi$Delay == "120",]
dataCi300 <- dataCi[dataCi$Delay == "300",]
m1 <- lm(dataCi0$Aapp ~ dataCi0$Cifull)
summary(m1)##
## Call:
## lm(formula = dataCi0$Aapp ~ dataCi0$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.13165 -0.04518 0.01711 0.05408 0.06699
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -5.2248105 0.0199353 -262.1 <2e-16 ***
## dataCi0$Cifull 0.1228632 0.0003089 397.8 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.06081 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.582e+05 on 1 and 69 DF, p-value: < 2.2e-16Gamma0 <- -m1$coefficients[1] / m1$coefficients[2]
m2 <- lm(dataCi15$Aapp ~ dataCi15$Cifull)
summary(m2)##
## Call:
## lm(formula = dataCi15$Aapp ~ dataCi15$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.13119 -0.04502 0.01705 0.05389 0.06676
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -5.0873326 0.0198665 -256.1 <2e-16 ***
## dataCi15$Cifull 0.1225900 0.0003078 398.3 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.0606 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.586e+05 on 1 and 69 DF, p-value: < 2.2e-16Gamma15 <- -m2$coefficients[1] / m2$coefficients[2]
m3 <- lm(dataCi30$Aapp ~ dataCi30$Cifull)
summary(m3)##
## Call:
## lm(formula = dataCi30$Aapp ~ dataCi30$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.13076 -0.04488 0.01699 0.05372 0.06654
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -4.9553180 0.0198028 -250.2 <2e-16 ***
## dataCi30$Cifull 0.1223321 0.0003068 398.7 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.0604 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.59e+05 on 1 and 69 DF, p-value: < 2.2e-16Gamma30 <- -m3$coefficients[1] / m3$coefficients[2]
m4 <- lm(dataCi60$Aapp ~ dataCi60$Cifull)
summary(m4)##
## Call:
## lm(formula = dataCi60$Aapp ~ dataCi60$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.13001 -0.04462 0.01690 0.05341 0.06616
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -4.706429 0.019689 -239.0 <2e-16 ***
## dataCi60$Cifull 0.121858 0.000305 399.5 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.06006 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.596e+05 on 1 and 69 DF, p-value: < 2.2e-16Gamma60 <- -m4$coefficients[1] / m4$coefficients[2]
m5 <- lm(dataCi120$Aapp ~ dataCi120$Cifull)
summary(m5)##
## Call:
## lm(formula = dataCi120$Aapp ~ dataCi120$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.12879 -0.04421 0.01673 0.05292 0.06555
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -4.2616980 0.0195062 -218.5 <2e-16 ***
## dataCi120$Cifull 0.1210486 0.0003022 400.5 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.0595 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.604e+05 on 1 and 69 DF, p-value: < 2.2e-16Gamma120 <- -m5$coefficients[1] / m5$coefficients[2]
m6 <- lm(dataCi300$Aapp ~ dataCi300$Cifull)
summary(m6)##
## Call:
## lm(formula = dataCi300$Aapp ~ dataCi300$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.12662 -0.04346 0.01645 0.05202 0.06444
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -3.2402030 0.0191773 -169.0 <2e-16 ***
## dataCi300$Cifull 0.1193852 0.0002971 401.8 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.05849 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.615e+05 on 1 and 69 DF, p-value: < 2.2e-16Gamma300 <- -m6$coefficients[1] / m6$coefficients[2]
GammastarCi <-
c(Gamma0, Gamma15, Gamma30, Gamma60, Gamma120, Gamma300)
# For Cc-based estimates, only use Cc < 75
dataCc <- data[data$Ccfull < 75,]
dataCc0 <- dataCc[dataCc$Delay == "0",]
dataCc15 <- dataCc[dataCc$Delay == "15",]
dataCc30 <- dataCc[dataCc$Delay == "30",]
dataCc60 <- dataCc[dataCc$Delay == "60",]
dataCc120 <- dataCc[dataCc$Delay == "120",]
dataCc300 <- dataCc[dataCc$Delay == "300",]
m1 <- lm(dataCc0$Aapp ~ dataCc0$Ccfull)
summary(m1)##
## Call:
## lm(formula = dataCc0$Aapp ~ dataCc0$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.075648 -0.025444 0.009857 0.031706 0.039431
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -6.4062011 0.0181852 -352.3 <2e-16 ***
## dataCc0$Ccfull 0.1438710 0.0003527 407.9 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03599 on 48 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 1.664e+05 on 1 and 48 DF, p-value: < 2.2e-16Gamma0 <- -m1$coefficients[1] / m1$coefficients[2]
m2 <- lm(dataCc15$Aapp ~ dataCc15$Ccfull)
summary(m2)##
## Call:
## lm(formula = dataCc15$Aapp ~ dataCc15$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.075393 -0.025359 0.009824 0.031599 0.039299
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -6.2659145 0.0181243 -345.7 <2e-16 ***
## dataCc15$Ccfull 0.1435470 0.0003515 408.4 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03587 on 48 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 1.668e+05 on 1 and 48 DF, p-value: < 2.2e-16Gamma15 <- -m2$coefficients[1] / m2$coefficients[2]
m3 <- lm(dataCc30$Aapp ~ dataCc30$Ccfull)
summary(m3)##
## Call:
## lm(formula = dataCc30$Aapp ~ dataCc30$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.075158 -0.025280 0.009794 0.031501 0.039177
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -6.1312578 0.0180681 -339.3 <2e-16 ***
## dataCc30$Ccfull 0.1432414 0.0003504 408.8 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03576 on 48 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 1.671e+05 on 1 and 48 DF, p-value: < 2.2e-16Gamma30 <- -m3$coefficients[1] / m3$coefficients[2]
m4 <- lm(dataCc60$Aapp ~ dataCc60$Ccfull)
summary(m4)##
## Call:
## lm(formula = dataCc60$Aapp ~ dataCc60$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.074737 -0.025139 0.009739 0.031325 0.038959
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -5.8775340 0.0179675 -327.1 <2e-16 ***
## dataCc60$Ccfull 0.1426797 0.0003485 409.4 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03556 on 48 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 1.676e+05 on 1 and 48 DF, p-value: < 2.2e-16Gamma60 <- -m4$coefficients[1] / m4$coefficients[2]
m5 <- lm(dataCc120$Aapp ~ dataCc120$Ccfull)
summary(m5)##
## Call:
## lm(formula = dataCc120$Aapp ~ dataCc120$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.074060 -0.024912 0.009651 0.031042 0.038607
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -5.4246412 0.0178052 -304.7 <2e-16 ***
## dataCc120$Ccfull 0.1417238 0.0003453 410.4 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03524 on 48 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 1.684e+05 on 1 and 48 DF, p-value: < 2.2e-16Gamma120 <- -m5$coefficients[1] / m5$coefficients[2]
m6 <- lm(dataCc300$Aapp ~ dataCc300$Ccfull)
summary(m6)##
## Call:
## lm(formula = dataCc300$Aapp ~ dataCc300$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.072835 -0.024500 0.009492 0.030529 0.037969
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -4.3867121 0.0175110 -250.5 <2e-16 ***
## dataCc300$Ccfull 0.1397661 0.0003396 411.5 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03466 on 48 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 1.694e+05 on 1 and 48 DF, p-value: < 2.2e-16Gamma300 <- -m6$coefficients[1] / m6$coefficients[2]
GammastarCc <-
c(Gamma0, Gamma15, Gamma30, Gamma60, Gamma120, Gamma300)
Delay2 <- c("0", "15", "30", "60", "120", "300")
PRcomps <-
as.data.frame(cbind(Delay2, GammaCc, GammaCi, GammastarCc, GammastarCi))
write.csv(PRcomps, "./data/PRcomps.csv")
ints <-
c(
m1$coefficients[1],
m2$coefficients[1],
m3$coefficients[1],
m4$coefficients[1],
m5$coefficients[1],
m6$coefficients[1]
)
dels <- c(0, 15, 30, 60, 120, 300)
summary(lm(ints ~ dels))##
## Call:
## lm(formula = ints ~ dels)
##
## Residuals:
## (Intercept) (Intercept).1 (Intercept).2 (Intercept).3 (Intercept).4
## -0.0751633 -0.0347043 0.0001247 0.0541933 0.1077758
## (Intercept).5
## -0.0522262
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -6.331038 0.041673 -151.92 1.13e-08 ***
## dels 0.006655 0.000309 21.54 2.75e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.07768 on 4 degrees of freedom
## Multiple R-squared: 0.9915, Adjusted R-squared: 0.9893
## F-statistic: 463.9 on 1 and 4 DF, p-value: 2.749e-05plot(ints ~ dels)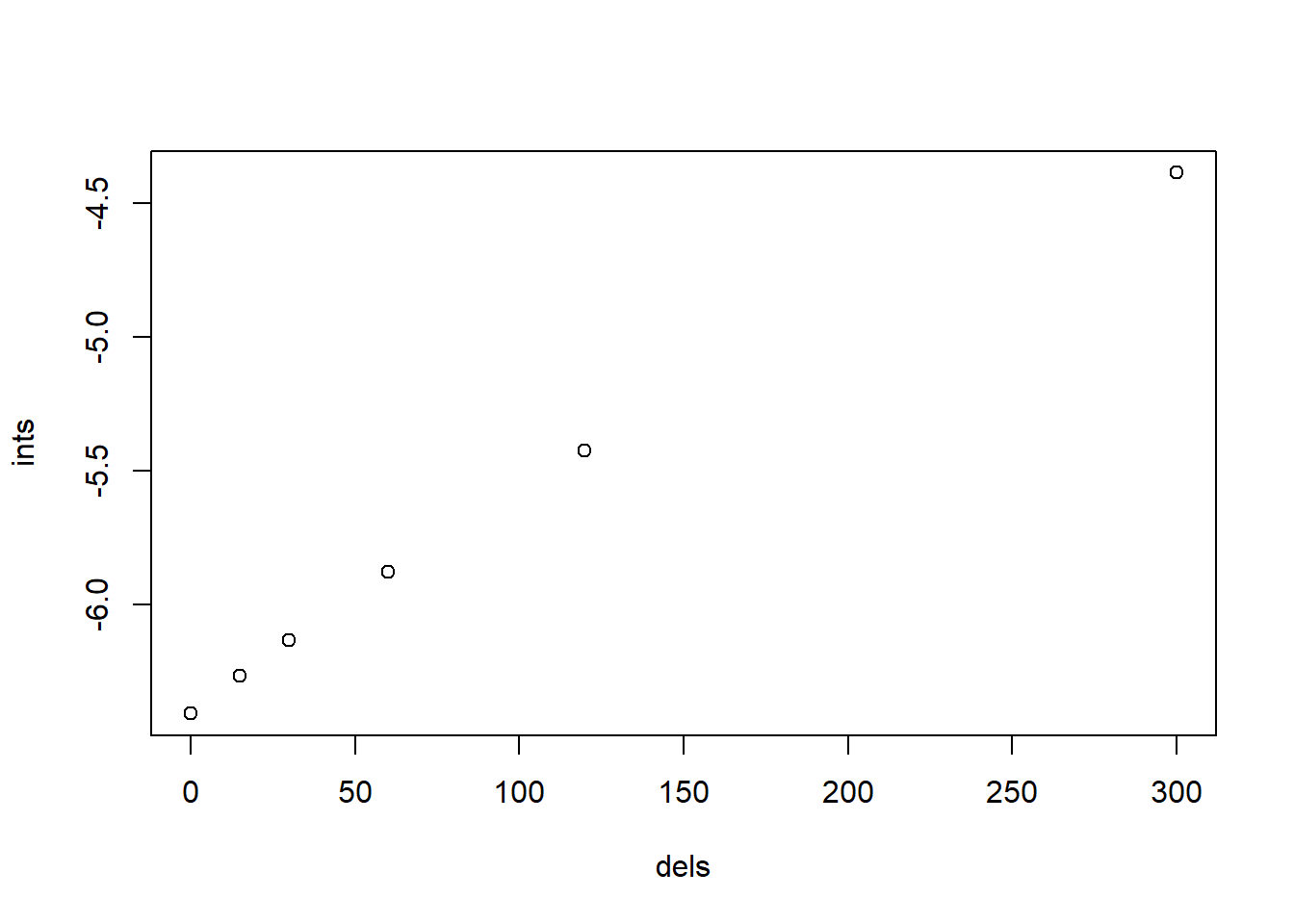
图 10.8: 基于 Cc 的不同延时下的截距
10.7.6 无光呼吸酶失活模块
该部分内容是在测量 ACi 曲线时检测 Rubisco 失活的影响 – 从激活状态的变化导致了多少的偏移?
10.7.6.1 数据构造
基于文献,假定 \(CO_2\) 从 400 ppm 降低至 5 ppm 时,激活率从 100% 降低至 80%。
#Assume that Rubisco activation state drops from 100% at 400 ppm to 80% at 5 ppm Cr (line 273 is Cr of 400, Cc of 297; 5ppm Cr is 25 ppm Cc) roughly from Salvucci et al 1986, arabidopsis; assume linear response
#不同的cr对应了不同的cc浓度,
#此为cc的变化范围(cr从400降低至5)
ccslope <- c(25, 297)
#酶的激活率变化
raslope <- c(0.80, 1.00)
#得到cc变化对应rubisco激活率变化的关系
ram1 <- lm(raslope ~ ccslope)
raslope <- coef(ram1)[2]
raint <- coef(ram1)[1]
#根据公式计算酶部分失活后各个参数
vora1 <- (raslope * Cc + raint) * Vomax * O2 / (O2 + Ko * (1 + Cc / Kc))
vcra1 <- (raslope * Cc + raint) * Vcmax * (Cc) / (Cc + Kco)
Ara1 <- vcra1 - 0.5 * vora1 - R
Aapparentra1 <- vcra1 - 0.5 * vora1
Cira1 <- Ara1 / gm + Cc
Cbra1 <- Ara1 / gsw + Cira1
Crra1 <- Ara1 / BLC + Cbra1
#失活后换算为分钟的变化斜率
Counter <- as.numeric(c(1:length(Cc)))
RateCr1model <- lm(Crra1 ~ Counter)
RateCr1 <- coef(RateCr1model)[2] * 60
RateCb1model <- lm(Cbra1 ~ Counter)
RateCb1 <- coef(RateCb1model)[2] * 60
RateCi1model <- lm(Cira1 ~ Counter)
RateCi1 <- coef(RateCi1model)[2] * 60
RateCcmodel <- lm(Cc ~ Counter)
RateCc <- coef(RateCcmodel)[2] * 60
#假定在5ppm时下降为40%
ccslope2 <- c(25, 297)
raslope2 <- c(0.40, 1.00)
ram2 <- lm(raslope2 ~ ccslope2)
raslope2 <- coef(ram2)[2]
raint2 <- coef(ram2)[1]
vora2 <-
(raslope2 * Cc + raint2) * Vomax * O2 / (O2 + Ko * (1 + Cc / Kc))
vcra2 <- (raslope2 * Cc + raint2) * Vcmax * (Cc) / (Cc + Kco)
Ara2 <- vcra2 - 0.5 * vora2 - R
Aapparentra2 <- vcra2 - 0.5 * vora2
Cira2 <- Ara2 / gm + Cc #umol mol-1
Cbra2 <- Ara2 / gsw + Cira2 #umol mol-1
Crra2 <- Ara2 / BLC + Cbra2 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCr2model <- lm(Crra2 ~ Counter)
RateCr2 <- coef(RateCr2model)[2] * 60 #umol mol-1 min-1
RateCb2model <- lm(Cbra2 ~ Counter)
RateCb2 <- coef(RateCb2model)[2] * 60 #umol mol-1 min-1
RateCi2model <- lm(Cira2 ~ Counter)
RateCi2 <- coef(RateCi2model)[2] * 60 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc <- coef(RateCcmodel)[2] * 60 #umol mol-1 min-1
#假定在5ppm时下降为20%
ccslope3 <- c(25, 297)
raslope3 <- c(0.20, 1.00)
ram3 <- lm(raslope3 ~ ccslope3)
raslope3 <- coef(ram3)[2]
raint3 <- coef(ram3)[1]
vora3 <-
(raslope3 * Cc + raint3) * Vomax * O2 / (O2 + Ko * (1 + Cc / Kc))
vcra3 <- (raslope3 * Cc + raint3) * Vcmax * (Cc) / (Cc + Kco)
Ara3 <- vcra3 - 0.5 * vora3 - R
Aapparentra3 <- vcra3 - 0.5 * vora3
Cira3 <- Ara3 / gm + Cc
Cbra3 <- Ara3 / gsw + Cira3
Crra3 <- Ara3 / BLC + Cbra3
Counter <- as.numeric(c(1:length(Cc)))
RateCr3model <- lm(Crra3 ~ Counter)
RateCr3 <- coef(RateCr3model)[2] * 60
RateCb3model <- lm(Cbra3 ~ Counter)
RateCb3 <- coef(RateCb3model)[2] * 60
RateCi3model <- lm(Cira3 ~ Counter)
RateCi3 <- coef(RateCi3model)[2] * 60
RateCcmodel <- lm(Cc ~ Counter)
RateCc <- coef(RateCcmodel)[2] * 60
Anet <- c(A, Ara1, Ara2, Ara3)
Aapp <- c(Aapparent, Aapparentra1, Aapparentra2, Aapparentra3)
Ccfull <- rep(Cc, 4)
Cifull <- c(Ci, Cira1, Cira2, Cira3)
Deactivation <-
c(
rep("None", length(A)),
rep("Low", length(Ara1)),
rep("Medium", length(Ara2)),
rep("High", length(Ara3))
)
RASdata <-
as.data.frame(cbind(Anet, Aapp, Ccfull, Cifull, Deactivation))
write.csv(RASdata, "./data/RASdata.csv")10.7.7 酶失活作图
data <- read.csv("./data/RASdata.csv")
data$Ccfull <- as.numeric(data$Ccfull)
data$Deactivation <- as.factor(data$Deactivation)
AnetCc <-
ggplot(data, aes(x = Ccfull, y = Anet, colour = Deactivation)) +
geom_point() +
labs(x = expression(C[c] ~ "(" * mu * mol ~ mol ^ {
-1
} * ")"),
y = expression(A[net] ~ "(" * mu * mol ~ m ^ {
-2
} ~ s ^ {
-1
} * ")")) +
labs(colour = 'Deactivation') +
scale_x_continuous(limits = c(25, 100),
breaks = c(25, 40, 55, 70, 85, 100)) +
scale_y_continuous(limits = c(-5, 5)) +
scale_colour_brewer(palette = 'Spectral') +
geom_hline(yintercept = 0) +
theme_bw() +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank())
AnetCc## Warning: Removed 1205 rows containing missing values (geom_point).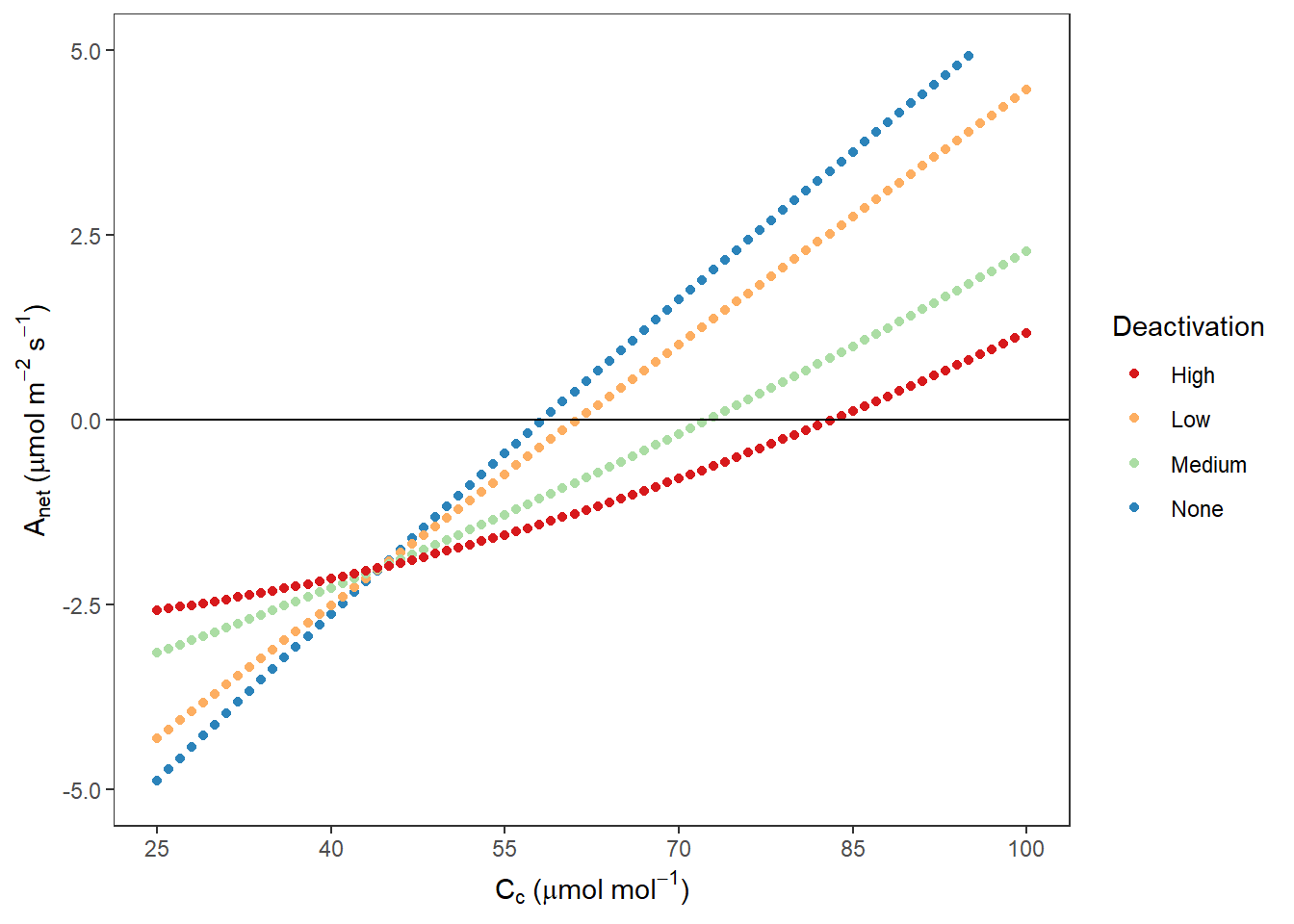
图 10.9: Rubisco 不同失活程度时 Anet VS Cc
AnetCi <-
ggplot(data, aes(x = Cifull, y = Anet, colour = Deactivation)) +
geom_point() +
labs(x = expression(C[i] ~ "(" * mu * mol ~ mol ^ {
-1
} * ")"),
y = expression(A[net] ~ "(" * mu * mol ~ m ^ {
-2
} ~ s ^ {
-1
} * ")")) +
labs(colour = 'Deactivation') +
scale_x_continuous(limits = c(25, 100),
breaks = c(25, 40, 55, 70, 85, 100)) +
scale_y_continuous(limits = c(-5, 5)) +
scale_colour_brewer(palette = 'Spectral') +
geom_hline(yintercept = 0) +
theme_bw() +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank())
AnetCi## Warning: Removed 1230 rows containing missing values (geom_point).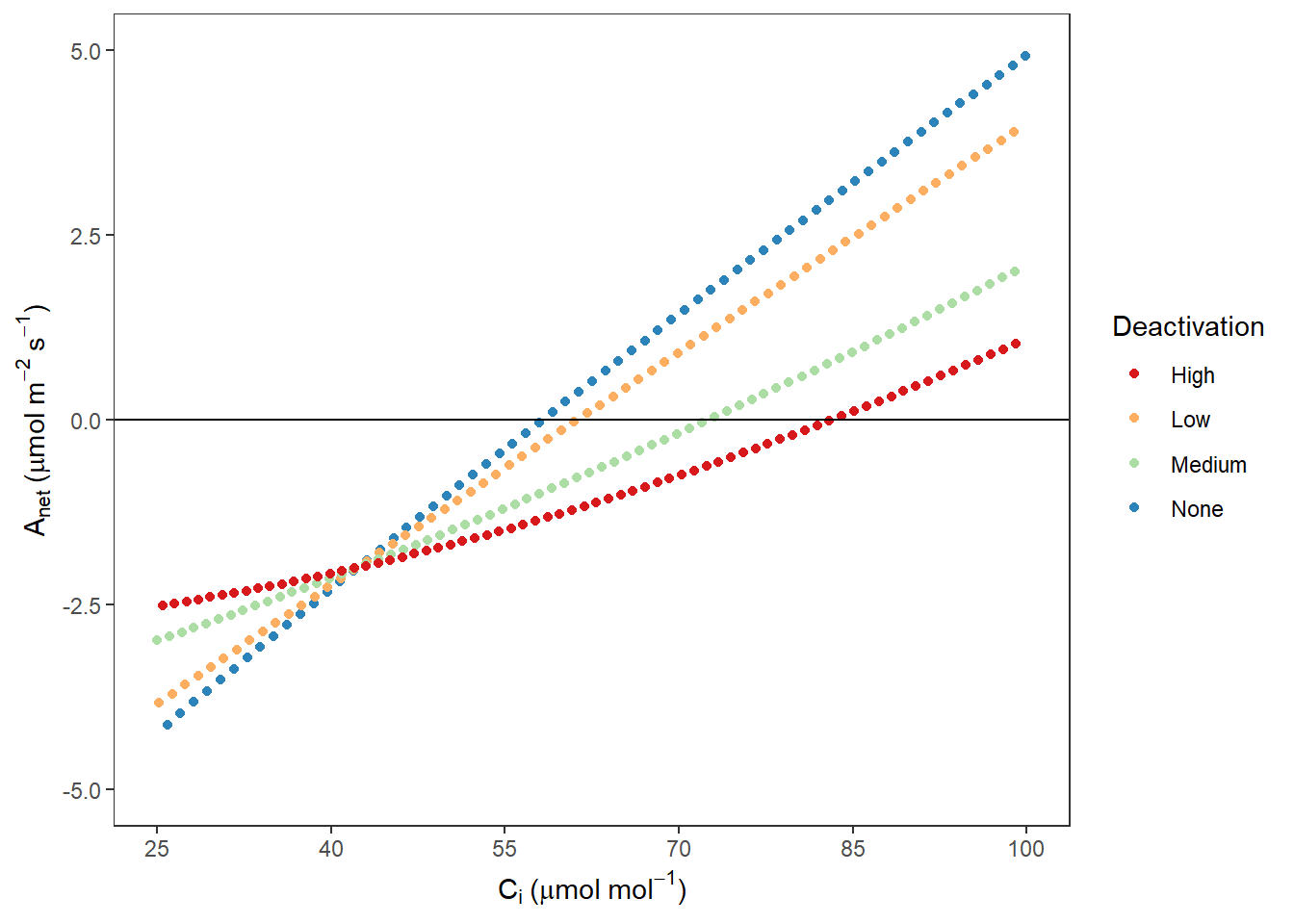
图 10.10: Rubisco 不同失活程度时 Anet VS Ci
AappCc <-
ggplot(data, aes(x = Ccfull, y = Aapp, colour = Deactivation)) +
geom_point() +
labs(x = expression(C[c] ~ "(" * mu * mol ~ mol ^ {
-1
} * ")"),
y = expression(A[apparent] ~ "(" * mu * mol ~ m ^ {
-2
} ~ s ^ {
-1
} * ")")) +
labs(colour = 'Deactivation') +
scale_x_continuous(limits = c(25, 75),
breaks = c(25, 35, 45, 55, 65, 75)) +
scale_y_continuous(limits = c(-5, 5)) +
scale_colour_brewer(palette = 'Spectral') +
geom_hline(yintercept = 0) +
theme_bw() +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank())
AappCc## Warning: Removed 1300 rows containing missing values (geom_point).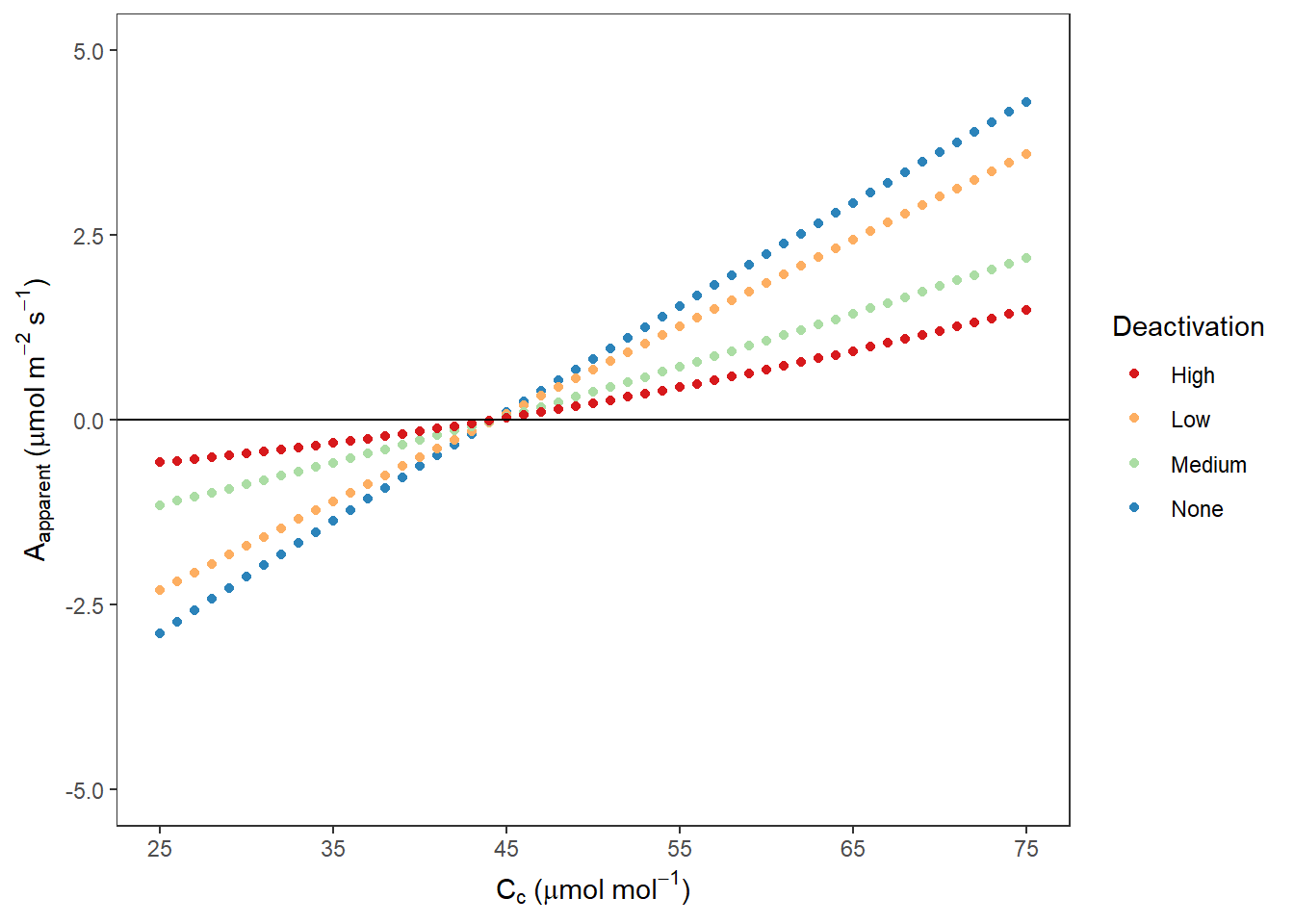
图 10.11: Rubisco 不同失活程度时 Aapp VS Cc
AappCi <-
ggplot(data, aes(x = Cifull, y = Aapp, colour = Deactivation)) +
geom_point() +
labs(x = expression(C[i] ~ "(" * mu * mol ~ mol ^ {
-1
} * ")"),
y = expression(A[apparent] ~ "(" * mu * mol ~ m ^ {
-2
} ~ s ^ {
-1
} * ")")) +
labs(colour = 'Deactivation') +
scale_x_continuous(limits = c(25, 75),
breaks = c(25, 35, 45, 55, 65, 75)) +
scale_y_continuous(limits = c(-5, 5)) +
scale_colour_brewer(palette = 'Spectral') +
geom_hline(yintercept = 0) +
theme_bw() +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank())
AappCi## Warning: Removed 1321 rows containing missing values (geom_point).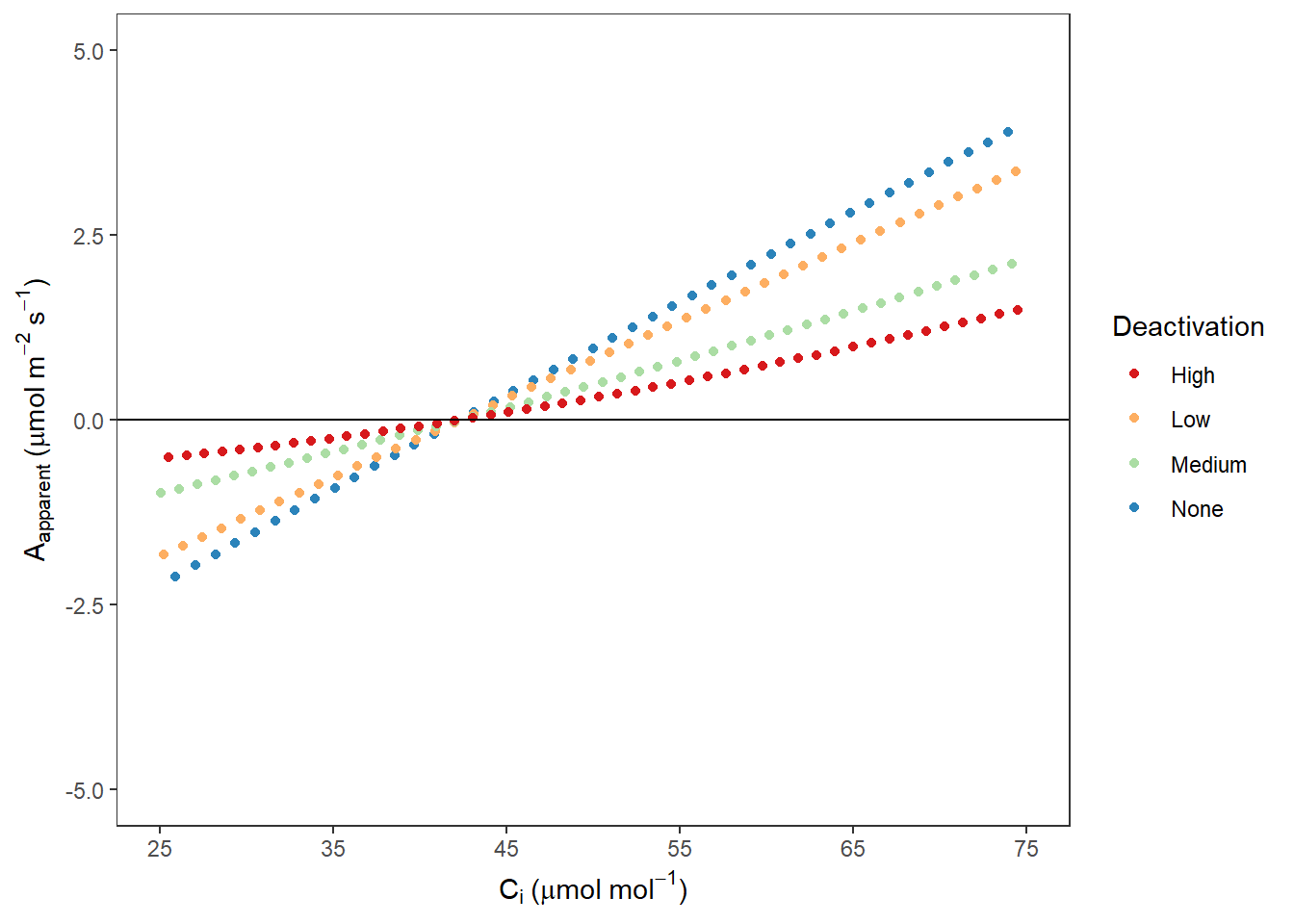
图 10.12: Rubisco 不同失活程度时 Aapp VS Ci
10.7.8 不同失活程度下补偿点计算
此部分内容同未失活状态相似,不在额外介绍,可参考 ?? 内容。
#Gamma
# For Ci-based estimates, only use Ci < 100
dataCi <- data[data$Cifull < 100, ]
dataCinone <- dataCi[dataCi$Deactivation == "None", ]
dataCilow <- dataCi[dataCi$Deactivation == "Low", ]
dataCimedium <- dataCi[dataCi$Deactivation == "Medium", ]
dataCihigh <- dataCi[dataCi$Deactivation == "High", ]
m1 <- lm(dataCinone$Anet ~ dataCinone$Cifull)
summary(m1)##
## Call:
## lm(formula = dataCinone$Anet ~ dataCinone$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.13165 -0.04518 0.01711 0.05408 0.06699
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -7.2248105 0.0199353 -362.4 <2e-16 ***
## dataCinone$Cifull 0.1228632 0.0003089 397.8 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.06081 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.582e+05 on 1 and 69 DF, p-value: < 2.2e-16Gammanone <- -m1$coefficients[1] / m1$coefficients[2]
m2 <- lm(dataCilow$Anet ~ dataCilow$Cifull)
summary(m2)##
## Call:
## lm(formula = dataCilow$Anet ~ dataCilow$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.034896 -0.011957 0.004499 0.014311 0.017726
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -6.444e+00 5.343e-03 -1206 <2e-16 ***
## dataCilow$Cifull 1.049e-01 8.341e-05 1258 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.01609 on 69 degrees of freedom
## Multiple R-squared: 1, Adjusted R-squared: 1
## F-statistic: 1.582e+06 on 1 and 69 DF, p-value: < 2.2e-16Gammalow <- -m2$coefficients[1] / m2$coefficients[2]
m3 <- lm(dataCimedium$Anet ~ dataCimedium$Cifull)
summary(m3)##
## Call:
## lm(formula = dataCimedium$Anet ~ dataCimedium$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.09168 -0.07338 -0.02281 0.05893 0.18150
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -4.8015337 0.0277371 -173.1 <2e-16 ***
## dataCimedium$Cifull 0.0671064 0.0004311 155.7 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.0832 on 71 degrees of freedom
## Multiple R-squared: 0.9971, Adjusted R-squared: 0.997
## F-statistic: 2.423e+04 on 1 and 71 DF, p-value: < 2.2e-16Gammamedium <- -m3$coefficients[1] / m3$coefficients[2]
m4 <- lm(dataCihigh$Anet ~ dataCihigh$Cifull)
summary(m4)##
## Call:
## lm(formula = dataCihigh$Anet ~ dataCihigh$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.15430 -0.12508 -0.03863 0.10156 0.30621
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -3.9450658 0.0467537 -84.38 <2e-16 ***
## dataCihigh$Cifull 0.0473551 0.0007255 65.27 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.1399 on 72 degrees of freedom
## Multiple R-squared: 0.9834, Adjusted R-squared: 0.9831
## F-statistic: 4260 on 1 and 72 DF, p-value: < 2.2e-16Gammahigh <- -m4$coefficients[1] / m4$coefficients[2]
GammaCi <- c(Gammanone, Gammalow, Gammamedium, Gammahigh)
# For Cc-based estimates, only use Cc < 75
dataCc <- data[data$Ccfull < 75, ]
dataCcnone <- dataCc[dataCc$Deactivation == "None", ]
dataCclow <- dataCc[dataCc$Deactivation == "Low", ]
dataCcmedium <- dataCc[dataCc$Deactivation == "Medium", ]
dataCchigh <- dataCc[dataCc$Deactivation == "High", ]
m1 <- lm(dataCcnone$Anet ~ dataCcnone$Ccfull)
summary(m1)##
## Call:
## lm(formula = dataCcnone$Anet ~ dataCcnone$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.075648 -0.025444 0.009857 0.031706 0.039431
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -8.4062011 0.0181852 -462.3 <2e-16 ***
## dataCcnone$Ccfull 0.1438710 0.0003527 407.9 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03599 on 48 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 1.664e+05 on 1 and 48 DF, p-value: < 2.2e-16Gammanone <- -m1$coefficients[1] / m1$coefficients[2]
m2 <- lm(dataCclow$Anet ~ dataCclow$Ccfull)
summary(m2)##
## Call:
## lm(formula = dataCclow$Anet ~ dataCclow$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.019617 -0.006598 0.002556 0.008222 0.010225
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -7.243e+00 4.716e-03 -1536 <2e-16 ***
## dataCclow$Ccfull 1.182e-01 9.146e-05 1292 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.009333 on 48 degrees of freedom
## Multiple R-squared: 1, Adjusted R-squared: 1
## F-statistic: 1.67e+06 on 1 and 48 DF, p-value: < 2.2e-16Gammalow <- -m2$coefficients[1] / m2$coefficients[2]
m3 <- lm(dataCcmedium$Anet ~ dataCcmedium$Ccfull)
summary(m3)##
## Call:
## lm(formula = dataCcmedium$Anet ~ dataCcmedium$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.04819 -0.03875 -0.01205 0.03109 0.09244
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -4.917283 0.022223 -221.3 <2e-16 ***
## dataCcmedium$Ccfull 0.066832 0.000431 155.1 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.04398 on 48 degrees of freedom
## Multiple R-squared: 0.998, Adjusted R-squared: 0.998
## F-statistic: 2.404e+04 on 1 and 48 DF, p-value: < 2.2e-16Gammamedium <- -m3$coefficients[1] / m3$coefficients[2]
m4 <- lm(dataCchigh$Anet ~ dataCchigh$Ccfull)
summary(m4)##
## Call:
## lm(formula = dataCchigh$Anet ~ dataCchigh$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.07739 -0.06223 -0.01935 0.04994 0.14847
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -3.7543099 0.0356922 -105.19 <2e-16 ***
## dataCchigh$Ccfull 0.0411528 0.0006922 59.45 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.07064 on 48 degrees of freedom
## Multiple R-squared: 0.9866, Adjusted R-squared: 0.9863
## F-statistic: 3534 on 1 and 48 DF, p-value: < 2.2e-16Gammahigh <- -m4$coefficients[1] / m4$coefficients[2]
GammaCc <- c(Gammanone, Gammalow, Gammamedium, Gammahigh)
#GammaStar
# For Ci-based estimates, only use Ci < 100
dataCi <- data[data$Cifull < 100, ]
dataCinone <- dataCi[dataCi$Deactivation == "None", ]
dataCilow <- dataCi[dataCi$Deactivation == "Low", ]
dataCimedium <- dataCi[dataCi$Deactivation == "Medium", ]
dataCihigh <- dataCi[dataCi$Deactivation == "High", ]
m1 <- lm(dataCinone$Aapp ~ dataCinone$Cifull)
summary(m1)##
## Call:
## lm(formula = dataCinone$Aapp ~ dataCinone$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.13165 -0.04518 0.01711 0.05408 0.06699
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -5.2248105 0.0199353 -262.1 <2e-16 ***
## dataCinone$Cifull 0.1228632 0.0003089 397.8 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.06081 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.582e+05 on 1 and 69 DF, p-value: < 2.2e-16Gammastarnone <- -m1$coefficients[1] / m1$coefficients[2]
m2 <- lm(dataCilow$Aapp ~ dataCilow$Cifull)
summary(m2)##
## Call:
## lm(formula = dataCilow$Aapp ~ dataCilow$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.034896 -0.011957 0.004499 0.014311 0.017726
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -4.444e+00 5.343e-03 -831.7 <2e-16 ***
## dataCilow$Cifull 1.049e-01 8.341e-05 1257.8 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.01609 on 69 degrees of freedom
## Multiple R-squared: 1, Adjusted R-squared: 1
## F-statistic: 1.582e+06 on 1 and 69 DF, p-value: < 2.2e-16Gammastarlow <- -m2$coefficients[1] / m2$coefficients[2]
m3 <- lm(dataCimedium$Aapp ~ dataCimedium$Cifull)
summary(m3)##
## Call:
## lm(formula = dataCimedium$Aapp ~ dataCimedium$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.09168 -0.07338 -0.02281 0.05893 0.18150
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -2.8015337 0.0277371 -101.0 <2e-16 ***
## dataCimedium$Cifull 0.0671064 0.0004311 155.7 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.0832 on 71 degrees of freedom
## Multiple R-squared: 0.9971, Adjusted R-squared: 0.997
## F-statistic: 2.423e+04 on 1 and 71 DF, p-value: < 2.2e-16Gammastarmedium <- -m3$coefficients[1] / m3$coefficients[2]
m4 <- lm(dataCihigh$Aapp ~ dataCihigh$Cifull)
summary(m4)##
## Call:
## lm(formula = dataCihigh$Aapp ~ dataCihigh$Cifull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.15430 -0.12508 -0.03863 0.10156 0.30621
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -1.9450658 0.0467537 -41.60 <2e-16 ***
## dataCihigh$Cifull 0.0473551 0.0007255 65.27 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.1399 on 72 degrees of freedom
## Multiple R-squared: 0.9834, Adjusted R-squared: 0.9831
## F-statistic: 4260 on 1 and 72 DF, p-value: < 2.2e-16Gammastarhigh <- -m4$coefficients[1] / m4$coefficients[2]
GammastarCi <-
c(Gammastarnone, Gammastarlow, Gammastarmedium, Gammastarhigh)
# For Cc-based estimates, only use Cc < 75
dataCc <- data[data$Ccfull < 75, ]
dataCcnone <- dataCc[dataCc$Deactivation == "None", ]
dataCclow <- dataCc[dataCc$Deactivation == "Low", ]
dataCcmedium <- dataCc[dataCc$Deactivation == "Medium", ]
dataCchigh <- dataCc[dataCc$Deactivation == "High", ]
m1 <- lm(dataCcnone$Aapp ~ dataCcnone$Ccfull)
summary(m1)##
## Call:
## lm(formula = dataCcnone$Aapp ~ dataCcnone$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.075648 -0.025444 0.009857 0.031706 0.039431
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -6.4062011 0.0181852 -352.3 <2e-16 ***
## dataCcnone$Ccfull 0.1438710 0.0003527 407.9 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03599 on 48 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 1.664e+05 on 1 and 48 DF, p-value: < 2.2e-16Gammastarnone <- -m1$coefficients[1] / m1$coefficients[2]
m2 <- lm(dataCclow$Aapp ~ dataCclow$Ccfull)
summary(m2)##
## Call:
## lm(formula = dataCclow$Aapp ~ dataCclow$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.019617 -0.006598 0.002556 0.008222 0.010225
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -5.243e+00 4.716e-03 -1112 <2e-16 ***
## dataCclow$Ccfull 1.182e-01 9.146e-05 1292 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.009333 on 48 degrees of freedom
## Multiple R-squared: 1, Adjusted R-squared: 1
## F-statistic: 1.67e+06 on 1 and 48 DF, p-value: < 2.2e-16Gammastarlow <- -m2$coefficients[1] / m2$coefficients[2]
m3 <- lm(dataCcmedium$Aapp ~ dataCcmedium$Ccfull)
summary(m3)##
## Call:
## lm(formula = dataCcmedium$Aapp ~ dataCcmedium$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.04819 -0.03875 -0.01205 0.03109 0.09244
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -2.917283 0.022223 -131.3 <2e-16 ***
## dataCcmedium$Ccfull 0.066832 0.000431 155.1 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.04398 on 48 degrees of freedom
## Multiple R-squared: 0.998, Adjusted R-squared: 0.998
## F-statistic: 2.404e+04 on 1 and 48 DF, p-value: < 2.2e-16Gammastarmedium <- -m3$coefficients[1] / m3$coefficients[2]
m4 <- lm(dataCchigh$Aapp ~ dataCchigh$Ccfull)
summary(m4)##
## Call:
## lm(formula = dataCchigh$Aapp ~ dataCchigh$Ccfull)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.07739 -0.06223 -0.01935 0.04994 0.14847
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -1.7543099 0.0356922 -49.15 <2e-16 ***
## dataCchigh$Ccfull 0.0411528 0.0006922 59.45 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.07064 on 48 degrees of freedom
## Multiple R-squared: 0.9866, Adjusted R-squared: 0.9863
## F-statistic: 3534 on 1 and 48 DF, p-value: < 2.2e-16Gammastarhigh <- -m4$coefficients[1] / m4$coefficients[2]
GammastarCc <-
c(Gammastarnone, Gammastarlow, Gammastarmedium, Gammastarhigh)
Deactivation2 <- c("None", "Low", "Medium", "High")
RAScomps <-
as.data.frame(cbind(Deactivation2, GammaCc, GammaCi, GammastarCc, GammastarCi))
write.csv(RAScomps, "./data/RAScomps.csv")10.8 时间延迟的扩散限制
对于扩散限制,下面的内容比较了多速率 RACiR 和标准 ACi 曲线的差别,比较实在有光呼吸和没有光呼吸的两种情况。对于没有扩散限制的表观光合速,采用了已知质量的碱石灰药品,放置于 1.7 ml 的微量离心管内,然后将其置于荧光叶室内部模拟叶片,此时叶室环境控制与其他实验不同,此时不再控制 H2OR。RACiR 测试从 500 到 0 的变化,不同样品的测量是随机的。
下面内容采用了一定的假设,来计算扩散的时间。
#Equations from Campbell & Norman, 1998
#We are taking a simple approach to calculating diffusion times.
#Here we make the simplifying assumption that diffusion is pure, planar
#Diffusion, such that:
#gtot = phat * D / deltaZ
#where gtot is total conductance, phat is molar density of air in mol /m^3,
#D is diffusion coefficient in m2/s, deltaZ is pathlength in m
#Since PV = NRT, N/V = P/RT
#T in K, R in J K-1 mol-1, P in Pa
#phat = Patm/(RT)
phat = 100000 / (8.314 * 298.15)
#We also assume a linear pathlength
#Note, if diffusion is nonlinear or nonplanar, it will affect the value determined
#for D from this equation.
#D = gtot * deltaZ / phat
#Diffusion time, t, varies with D and deltaZ such that:
#t = (deltaZ)^2 / D
#So
#t = (deltaZ)^2 / (gtot * deltaZ / phat)
#If we assume mean diffusion pathlength of 1/2 lamina thickness,
#then Onoda et al. 2011 lamina thicknesses of: median 0.22 mm (0.11 to 0.74 for 95% CI)
#becomes 0.11 mm (0.055 to 0.37) for estimated deltaZ
#Convert pathlength to m
dZlow <- 0.055 / 1000
dZmedian <- 0.11 / 1000
dZhigh <- 0.37 / 1000下面的内容是对边界层导度和气孔导度等赋值,由此而计算出其他所需要的参数:
#Mesophyll Conductance
BLC <- 2 #mol m-2 s-1
gsw <- 0.4 #mol m-2 s-1
# 无限制的叶肉导度,并以此计算ci等
gm1 <- 1 #mol m-2 s-1
Ci1 <- A / gm1 + Cc #umol mol-1
Cim1 <- Ci1
Cb1 <- A / gsw + Ci1 #umol mol-1
Cr1 <- A / BLC + Cb1 #umol mol-1
#根据斜率计算达到 100 ppm min-1 时记录数据的个数
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr1 ~ Counter)
x1 <- 100 / coef(RateCrmodel)[2]
RateCr1 <- coef(RateCrmodel)[2] * x1 #umol mol-1 min-1
#计算ci,Cc等达到100ppm min-1 时数据的个数
RateCbmodel <- lm(Cb1 ~ Counter)
RateCb1 <- coef(RateCbmodel)[2] * x1 #umol mol-1 min-1
RateCimodel <- lm(Ci1 ~ Counter)
RateCi1 <- coef(RateCimodel)[2] * x1 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc1 <- coef(RateCcmodel)[2] * x1 #umol mol-1 min-1
#总的阻力
res1 <- 1 / gm1 + 1 / BLC + 1 / gsw
#不同的叶肉导度计算其他参数
gm2 <- 2 #mol m-2 s-1
Ci2 <- A / gm2 + Cc #umol mol-1
Cim2 <- Ci2
Cb2 <- A / gsw + Ci2 #umol mol-1
Cr2 <- A / BLC + Cb2 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr2 ~ Counter)
x2 <- 100 / coef(RateCrmodel)[2]
RateCr2 <- coef(RateCrmodel)[2] * x2 #umol mol-1 min-1
RateCbmodel <- lm(Cb2 ~ Counter)
RateCb2 <- coef(RateCbmodel)[2] * x2 #umol mol-1 min-1
RateCimodel <- lm(Ci2 ~ Counter)
RateCi2 <- coef(RateCimodel)[2] * x2 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc2 <- coef(RateCcmodel)[2] * x2 #umol mol-1 min-1
res2 <- 1 / gm2 + 1 / BLC + 1 / gsw
#再次计算不同导度下的数值
gm4 <- 4 #mol m-2 s-1
Ci4 <- A / gm4 + Cc #umol mol-1
Cim4 <- Ci4
Cb4 <- A / gsw + Ci4 #umol mol-1
Cr4 <- A / BLC + Cb4 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr4 ~ Counter)
x4 <- 100 / coef(RateCrmodel)[2]
RateCr4 <- coef(RateCrmodel)[2] * x4 #umol mol-1 min-1
RateCbmodel <- lm(Cb4 ~ Counter)
RateCb4 <- coef(RateCbmodel)[2] * x4 #umol mol-1 min-1
RateCimodel <- lm(Ci4 ~ Counter)
RateCi4 <- coef(RateCimodel)[2] * x4 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc4 <- coef(RateCcmodel)[2] * x4 #umol mol-1 min-1
res4 <- 1 / gm4 + 1 / BLC + 1 / gsw
#再次计算不同导度下的数值
gm05 <- 0.5 #mol m-2 s-1
Ci05 <- A / gm05 + Cc #umol mol-1
Cim05 <- Ci05
Cb05 <- A / gsw + Ci05 #umol mol-1
Cr05 <- A / BLC + Cb05 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr05 ~ Counter)
x05 <- 100 / coef(RateCrmodel)[2]
RateCr05 <- coef(RateCrmodel)[2] * x05 #umol mol-1 min-1
RateCbmodel <- lm(Cb05 ~ Counter)
RateCb05 <- coef(RateCbmodel)[2] * x05 #umol mol-1 min-1
RateCimodel <- lm(Ci05 ~ Counter)
RateCi05 <- coef(RateCimodel)[2] * x05 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc05 <- coef(RateCcmodel)[2] * x05 #umol mol-1 min-1
res05 <- 1 / gm05 + 1 / BLC + 1 / gsw
#正常的叶肉导度数据计算其他参数
gm025 <- 0.25 #mol m-2 s-1
Ci025 <- A / gm025 + Cc #umol mol-1
Cim025 <- Ci025
Cb025 <- A / gsw + Ci025 #umol mol-1
Cr025 <- A / BLC + Cb025 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr025 ~ Counter)
x025 <- 100 / coef(RateCrmodel)[2]
RateCr025 <- coef(RateCrmodel)[2] * x025 #umol mol-1 min-1
RateCbmodel <- lm(Cb025 ~ Counter)
RateCb025 <- coef(RateCbmodel)[2] * x025 #umol mol-1 min-1
RateCimodel <- lm(Ci025 ~ Counter)
RateCi025 <- coef(RateCimodel)[2] * x025 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc025 <- coef(RateCcmodel)[2] * x025 #umol mol-1 min-1
res025 <- 1 / gm025 + 1 / BLC + 1 / gsw
#另一个正常的叶肉导度
gm0125 <- 0.125 #mol m-2 s-1
Ci0125 <- A / gm0125 + Cc #umol mol-1
Cim0125 <- Ci0125
Cb0125 <- A / gsw + Ci0125 #umol mol-1
Cr0125 <- A / BLC + Cb0125 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr0125 ~ Counter)
x0125 <- 100 / coef(RateCrmodel)[2]
RateCr0125 <- coef(RateCrmodel)[2] * x0125 #umol mol-1 min-1
RateCbmodel <- lm(Cb0125 ~ Counter)
RateCb0125 <- coef(RateCbmodel)[2] * x0125 #umol mol-1 min-1
RateCimodel <- lm(Ci0125 ~ Counter)
RateCi0125 <- coef(RateCimodel)[2] * x0125 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc0125 <- coef(RateCcmodel)[2] * x0125 #umol mol-1 min-1
res0125 <- 1 / gm0125 + 1 / BLC + 1 / gsw
#利用不同叶肉导度的数据计算结果构造数据
Ratesgm <-
c(RateCc0125, RateCc025, RateCc05, RateCc1, RateCc2, RateCc4)
gmval <- c(0.125, 0.25, 0.5, 1, 2, 4)
totalresgm <- c(res0125, res025, res05, res1, res2, res4)
resistance <-
c(
rep(res0125, 376),
rep(res025, 376),
rep(res05, 376),
rep(res1, 376),
rep(res2, 376),
rep(res4, 376)
)
#其余部分与上面类似
#此时采用不同的气孔导度构建数据
BLC <- 2 #mol m-2 s-1
gm <- 1 #mol m-2 s-1
gsw <- 0.4 #mol m-2 s-1
Ci1 <- A / gm + Cc #umol mol-1
Cis04 <- Ci1
Cb1 <- A / gsw + Ci1 #umol mol-1
Cr1 <- A / BLC + Cb1 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr1 ~ Counter)
x1 <- 100 / coef(RateCrmodel)[2]
RateCr1 <- coef(RateCrmodel)[2] * x1 #umol mol-1 min-1
RateCbmodel <- lm(Cb1 ~ Counter)
RateCb1 <- coef(RateCbmodel)[2] * x1 #umol mol-1 min-1
RateCimodel <- lm(Ci1 ~ Counter)
RateCi1 <- coef(RateCimodel)[2] * x1 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc04 <- coef(RateCcmodel)[2] * x1 #umol mol-1 min-1
res04 <- 1 / gm + 1 / BLC + 1 / gsw
gsw <- 0.2 #mol m-2 s-1
Ci1 <- A / gm + Cc #umol mol-1
Cis02 <- Ci1
Cb1 <- A / gsw + Ci1 #umol mol-1
Cr1 <- A / BLC + Cb1 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr1 ~ Counter)
x1 <- 100 / coef(RateCrmodel)[2]
RateCr1 <- coef(RateCrmodel)[2] * x1 #umol mol-1 min-1
RateCbmodel <- lm(Cb1 ~ Counter)
RateCb1 <- coef(RateCbmodel)[2] * x1 #umol mol-1 min-1
RateCimodel <- lm(Ci1 ~ Counter)
RateCi1 <- coef(RateCimodel)[2] * x1 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc02 <- coef(RateCcmodel)[2] * x1 #umol mol-1 min-1
res02 <- 1 / gm + 1 / BLC + 1 / gsw
gsw <- 0.1 #mol m-2 s-1
Ci1 <- A / gm + Cc #umol mol-1
Cis01 <- Ci1
Cb1 <- A / gsw + Ci1 #umol mol-1
Cr1 <- A / BLC + Cb1 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr1 ~ Counter)
x1 <- 100 / coef(RateCrmodel)[2]
RateCr1 <- coef(RateCrmodel)[2] * x1 #umol mol-1 min-1
RateCbmodel <- lm(Cb1 ~ Counter)
RateCb1 <- coef(RateCbmodel)[2] * x1 #umol mol-1 min-1
RateCimodel <- lm(Ci1 ~ Counter)
RateCi1 <- coef(RateCimodel)[2] * x1 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc01 <- coef(RateCcmodel)[2] * x1 #umol mol-1 min-1
res01 <- 1 / gm + 1 / BLC + 1 / gsw
gsw <- 0.05 #mol m-2 s-1
Ci1 <- A / gm + Cc #umol mol-1
Cis05 <- Ci1
Cb1 <- A / gsw + Ci1 #umol mol-1
Cr1 <- A / BLC + Cb1 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr1 ~ Counter)
x1 <- 100 / coef(RateCrmodel)[2]
RateCr1 <- coef(RateCrmodel)[2] * x1 #umol mol-1 min-1
RateCbmodel <- lm(Cb1 ~ Counter)
RateCb1 <- coef(RateCbmodel)[2] * x1 #umol mol-1 min-1
RateCimodel <- lm(Ci1 ~ Counter)
RateCi1 <- coef(RateCimodel)[2] * x1 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc005 <- coef(RateCcmodel)[2] * x1 #umol mol-1 min-1
res005 <- 1 / gm + 1 / BLC + 1 / gsw
gsw <- 0.025 #mol m-2 s-1
Ci1 <- A / gm + Cc #umol mol-1
Cis0025 <- Ci1
Cb1 <- A / gsw + Ci1 #umol mol-1
Cr1 <- A / BLC + Cb1 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr1 ~ Counter)
x1 <- 100 / coef(RateCrmodel)[2]
RateCr1 <- coef(RateCrmodel)[2] * x1 #umol mol-1 min-1
RateCbmodel <- lm(Cb1 ~ Counter)
RateCb1 <- coef(RateCbmodel)[2] * x1 #umol mol-1 min-1
RateCimodel <- lm(Ci1 ~ Counter)
RateCi1 <- coef(RateCimodel)[2] * x1 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc0025 <- coef(RateCcmodel)[2] * x1 #umol mol-1 min-1
res0025 <- 1 / gm + 1 / BLC + 1 / gsw
Ratesgsw <- c(RateCc0025, RateCc005, RateCc01, RateCc02, RateCc04)
gswvals <- c(0.025, 0.05, 0.1, 0.2, 0.4)
totalresgsw <- c(res0025, res005, res01, res02, res04)
# 下面的代码是采用不同的边界层导度
# 含义与上面代码相似
gm <- 1 #mol m-2 s-1
gsw <- 0.4 #mol m-2 s-1
BLC <- 2 #mol m-2 s-1
Ci1 <- A / gm + Cc #umol mol-1
Cib2 <- Ci1
Cb1 <- A / gsw + Ci1 #umol mol-1
Cr1 <- A / BLC + Cb1 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr1 ~ Counter)
x1 <- 100 / coef(RateCrmodel)[2]
RateCr1 <- coef(RateCrmodel)[2] * x1 #umol mol-1 min-1
RateCbmodel <- lm(Cb1 ~ Counter)
RateCb1 <- coef(RateCbmodel)[2] * x1 #umol mol-1 min-1
RateCimodel <- lm(Ci1 ~ Counter)
RateCi1 <- coef(RateCimodel)[2] * x1 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc2 <- coef(RateCcmodel)[2] * x1 #umol mol-1 min-1
res2 <- 1 / gm + 1 / BLC + 1 / gsw
BLC <- 4 #mol m-2 s-1
Ci1 <- A / gm + Cc #umol mol-1
Cib4 <- Ci1
Cb1 <- A / gsw + Ci1 #umol mol-1
Cr1 <- A / BLC + Cb1 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr1 ~ Counter)
x1 <- 100 / coef(RateCrmodel)[2]
RateCr1 <- coef(RateCrmodel)[2] * x1 #umol mol-1 min-1
RateCbmodel <- lm(Cb1 ~ Counter)
RateCb1 <- coef(RateCbmodel)[2] * x1 #umol mol-1 min-1
RateCimodel <- lm(Ci1 ~ Counter)
RateCi1 <- coef(RateCimodel)[2] * x1 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc4 <- coef(RateCcmodel)[2] * x1 #umol mol-1 min-1
res4 <- 1 / gm + 1 / BLC + 1 / gsw
BLC <- 1 #mol m-2 s-1
Ci1 <- A / gm + Cc #umol mol-1
Cib1 <- Ci1
Cb1 <- A / gsw + Ci1 #umol mol-1
Cr1 <- A / BLC + Cb1 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr1 ~ Counter)
x1 <- 100 / coef(RateCrmodel)[2]
RateCr1 <- coef(RateCrmodel)[2] * x1 #umol mol-1 min-1
RateCbmodel <- lm(Cb1 ~ Counter)
RateCb1 <- coef(RateCbmodel)[2] * x1 #umol mol-1 min-1
RateCimodel <- lm(Ci1 ~ Counter)
RateCi1 <- coef(RateCimodel)[2] * x1 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc1 <- coef(RateCcmodel)[2] * x1 #umol mol-1 min-1
res1 <- 1 / gm + 1 / BLC + 1 / gsw
BLC <- 0.5 #mol m-2 s-1
Ci1 <- A / gm + Cc #umol mol-1
Cb1 <- A / gsw + Ci1 #umol mol-1
Cib05 <- Ci1
Cr1 <- A / BLC + Cb1 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr1 ~ Counter)
x1 <- 100 / coef(RateCrmodel)[2]
RateCr1 <- coef(RateCrmodel)[2] * x1 #umol mol-1 min-1
RateCbmodel <- lm(Cb1 ~ Counter)
RateCb1 <- coef(RateCbmodel)[2] * x1 #umol mol-1 min-1
RateCimodel <- lm(Ci1 ~ Counter)
RateCi1 <- coef(RateCimodel)[2] * x1 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc05 <- coef(RateCcmodel)[2] * x1 #umol mol-1 min-1
res05 <- 1 / gm + 1 / BLC + 1 / gsw
BLC <- 0.25 #mol m-2 s-1
Ci1 <- A / gm + Cc #umol mol-1
Cb1 <- A / gsw + Ci1 #umol mol-1
Cib025 <- Ci1
Cr1 <- A / BLC + Cb1 #umol mol-1
Counter <- as.numeric(c(1:length(Cc)))
RateCrmodel <- lm(Cr1 ~ Counter)
x1 <- 100 / coef(RateCrmodel)[2]
RateCr1 <- coef(RateCrmodel)[2] * x1 #umol mol-1 min-1
RateCbmodel <- lm(Cb1 ~ Counter)
RateCb1 <- coef(RateCbmodel)[2] * x1 #umol mol-1 min-1
RateCimodel <- lm(Ci1 ~ Counter)
RateCi1 <- coef(RateCimodel)[2] * x1 #umol mol-1 min-1
RateCcmodel <- lm(Cc ~ Counter)
RateCc025 <- coef(RateCcmodel)[2] * x1 #umol mol-1 min-1
res025 <- 1 / gm + 1 / BLC + 1 / gsw
BLCRates <- c(RateCc025, RateCc05, RateCc1, RateCc2, RateCc4)
BLCvals <- c(0.25, 0.5, 1, 2, 4)
totalresBLC <- c(res025, res05, res1, res2, res4)
Scenario <-
c(
rep("Boundary Layer Conductance", 5),
rep("Stomatal Conductance", 5),
rep("Mesophyll Conductance", 6)
)
Rates <- c(BLCRates, Ratesgsw, Ratesgm)
Conductances <- c(BLCvals, gswvals, gmval)
TotalRes <- c(totalresBLC, totalresgsw, totalresgm)
Cidiffusion <- c(Cim0125, Cim025, Cim05, Cim1, Cim2, Cim4)
Adiffusion <- rep(A, 6)
Aappdiffusion <- rep(Aapparent, 6)
variable <- c(rep("Mesophyll Conductance", 6 * length(Cim1)))
conductance <-
c(rep(0.125, 376),
rep(0.25, 376),
rep(0.5, 376),
rep(1, 376),
rep(2, 376),
rep(4, 376))
Diffusionplot <-
as.data.frame(cbind(
Cidiffusion,
Adiffusion,
Aappdiffusion,
variable,
conductance,
resistance
))
write.csv(Diffusionplot, "./data/DiffusionLimitsACI.csv")
Diffusion <-
as.data.frame(cbind(Scenario, Rates, Conductances, TotalRes))
write.csv(Diffusion, "./data/DiffusionLimits.csv")knitr::kable(head(Diffusion))| Scenario | Rates | Conductances | TotalRes | |
|---|---|---|---|---|
| Counter | Boundary Layer Conductance | 57.1493939574751 | 0.25 | 7.5 |
| Counter.1 | Boundary Layer Conductance | 64.522239461912 | 0.5 | 5.5 |
| Counter.2 | Boundary Layer Conductance | 68.9712299728077 | 1 | 4.5 |
| Counter.3 | Boundary Layer Conductance | 71.4340185663793 | 2 | 4 |
| Counter.4 | Boundary Layer Conductance | 72.7325667866589 | 4 | 3.75 |
| Counter.5 | Stomatal Conductance | 19.4216526694884 | 0.025 | 41.5 |
10.8.1 扩散限制滞后性
下面的代码,是根据上面代码的计算结果,结合最初的扩散时间的公式,来计算出各个参数的最大最小值,中间值,构造数据:
gtot1 <- 1 / res1
t1low = (dZlow) ^ 2 / (gtot1 * dZlow / phat)
t1median = (dZmedian) ^ 2 / (gtot1 * dZmedian / phat)
t1high = (dZhigh) ^ 2 / (gtot1 * dZhigh / phat)
Cc1low <- c(Cc + (t1low * RateCc1))
Cc1median <- c(Cc + (t1median * RateCc1))
Cc1high <- c(Cc + (t1high * RateCc1))
vo1low <- Vomax * O2 / (O2 + Ko * (1 + Cc1low / Kc))
A1low <- vc - 0.5 * vo1low - R
vo1median <- Vomax * O2 / (O2 + Ko * (1 + Cc1median / Kc))
A1median <- vc - 0.5 * vo1median - R
vo1high <- Vomax * O2 / (O2 + Ko * (1 + Cc1high / Kc))
A1high <- vc - 0.5 * vo1high - R
gtot0125 <- 1 / res0125
t0125low = (dZlow) ^ 2 / (gtot0125 * dZlow / phat)
t0125median = (dZmedian) ^ 2 / (gtot0125 * dZmedian / phat)
t0125high = (dZhigh) ^ 2 / (gtot0125 * dZhigh / phat)
Cc0125low <- c(Cc + (t0125low * RateCc0125))
Cc0125median <- c(Cc + (t0125median * RateCc0125))
Cc0125high <- c(Cc + (t0125high * RateCc0125))
vo0125low <- Vomax * O2 / (O2 + Ko * (1 + Cc0125low / Kc))
A0125low <- vc - 0.5 * vo0125low - R
vo0125median <- Vomax * O2 / (O2 + Ko * (1 + Cc0125median / Kc))
A0125median <- vc - 0.5 * vo0125median - R
vo0125high <- Vomax * O2 / (O2 + Ko * (1 + Cc0125high / Kc))
A0125high <- vc - 0.5 * vo0125high - R
gtot025 <- 1 / res025
t025low = (dZlow) ^ 2 / (gtot025 * dZlow / phat)
t025median = (dZmedian) ^ 2 / (gtot025 * dZmedian / phat)
t025high = (dZhigh) ^ 2 / (gtot025 * dZhigh / phat)
Cc025low <- c(Cc + (t025low * RateCc025))
Cc025median <- c(Cc + (t025median * RateCc025))
Cc025high <- c(Cc + (t025high * RateCc025))
vo025low <- Vomax * O2 / (O2 + Ko * (1 + Cc025low / Kc))
A025low <- vc - 0.5 * vo025low - R
vo025median <- Vomax * O2 / (O2 + Ko * (1 + Cc025median / Kc))
A025median <- vc - 0.5 * vo025median - R
vo025high <- Vomax * O2 / (O2 + Ko * (1 + Cc025high / Kc))
A025high <- vc - 0.5 * vo025high - R
gtot05 <- 1 / res05
t05low = (dZlow) ^ 2 / (gtot05 * dZlow / phat)
t05median = (dZmedian) ^ 2 / (gtot05 * dZmedian / phat)
t05high = (dZhigh) ^ 2 / (gtot05 * dZhigh / phat)
Cc05low <- c(Cc + (t05low * RateCc05))
Cc05median <- c(Cc + (t05median * RateCc05))
Cc05high <- c(Cc + (t05high * RateCc05))
vo05low <- Vomax * O2 / (O2 + Ko * (1 + Cc05low / Kc))
A05low <- vc - 0.5 * vo05low - R
vo05median <- Vomax * O2 / (O2 + Ko * (1 + Cc05median / Kc))
A05median <- vc - 0.5 * vo05median - R
vo05high <- Vomax * O2 / (O2 + Ko * (1 + Cc05high / Kc))
A05high <- vc - 0.5 * vo05high - R
gtot2 <- 1 / res2
t2low = (dZlow) ^ 2 / (gtot2 * dZlow / phat)
t2median = (dZmedian) ^ 2 / (gtot2 * dZmedian / phat)
t2high = (dZhigh) ^ 2 / (gtot2 * dZhigh / phat)
Cc2low <- c(Cc + (t2low * RateCc2))
Cc2median <- c(Cc + (t2median * RateCc2))
Cc2high <- c(Cc + (t2high * RateCc2))
vo2low <- Vomax * O2 / (O2 + Ko * (1 + Cc2low / Kc))
A2low <- vc - 0.5 * vo2low - R
vo2median <- Vomax * O2 / (O2 + Ko * (1 + Cc2median / Kc))
A2median <- vc - 0.5 * vo2median - R
vo2high <- Vomax * O2 / (O2 + Ko * (1 + Cc2high / Kc))
A2high <- vc - 0.5 * vo2high - R
gtot4 <- 1 / res4
t4low = (dZlow) ^ 2 / (gtot4 * dZlow / phat)
t4median = (dZmedian) ^ 2 / (gtot4 * dZmedian / phat)
t4high = (dZhigh) ^ 2 / (gtot4 * dZhigh / phat)
Cc4low <- c(Cc + (t4low * RateCc4))
Cc4median <- c(Cc + (t4median * RateCc4))
Cc4high <- c(Cc + (t4high * RateCc4))
vo4low <- Vomax * O2 / (O2 + Ko * (1 + Cc4low / Kc))
A4low <- vc - 0.5 * vo4low - R
vo4median <- Vomax * O2 / (O2 + Ko * (1 + Cc4median / Kc))
A4median <- vc - 0.5 * vo4median - R
vo4high <- Vomax * O2 / (O2 + Ko * (1 + Cc4high / Kc))
A4high <- vc - 0.5 * vo4high - R
Cc0125high2 <- c(Cc + (t0125high * RateCc0125 * 2))
Cc025high2 <- c(Cc + (t025high * RateCc025 * 2))
Cc05high2 <- c(Cc + (t05high * RateCc05 * 2))
Cc1high2 <- c(Cc + (t1high * RateCc1 * 2))
Cc2high2 <- c(Cc + (t2high * RateCc2 * 2))
Cc4high2 <- c(Cc + (t4high * RateCc4 * 2))
vo0125high2 <- Vomax * O2 / (O2 + Ko * (1 + Cc0125high2 / Kc))
A0125high2 <- vc - 0.5 * vo0125high2 - R
vo025high2 <- Vomax * O2 / (O2 + Ko * (1 + Cc025high2 / Kc))
A025high2 <- vc - 0.5 * vo025high2 - R
vo05high2 <- Vomax * O2 / (O2 + Ko * (1 + Cc05high2 / Kc))
A05high2 <- vc - 0.5 * vo05high2 - R
vo1high2 <- Vomax * O2 / (O2 + Ko * (1 + Cc1high2 / Kc))
A1high2 <- vc - 0.5 * vo1high2 - R
vo2high2 <- Vomax * O2 / (O2 + Ko * (1 + Cc2high2 / Kc))
A2high2 <- vc - 0.5 * vo2high2 - R
vo4high2 <- Vomax * O2 / (O2 + Ko * (1 + Cc4high2 / Kc))
A4high2 <- vc - 0.5 * vo4high2 - R
Ahigh2 <-
c(A0125high2, A025high2, A05high2, A1high2, A2high2, A4high2)
Cc0125high3 <- c(Cc + (t0125high * RateCc0125 * 3))
Cc025high3 <- c(Cc + (t025high * RateCc025 * 3))
Cc05high3 <- c(Cc + (t05high * RateCc05 * 3))
Cc1high3 <- c(Cc + (t1high * RateCc1 * 3))
Cc2high3 <- c(Cc + (t2high * RateCc2 * 3))
Cc4high3 <- c(Cc + (t4high * RateCc4 * 3))
vo0125high3 <- Vomax * O2 / (O2 + Ko * (1 + Cc0125high3 / Kc))
A0125high3 <- vc - 0.5 * vo0125high3 - R
vo025high3 <- Vomax * O2 / (O2 + Ko * (1 + Cc025high3 / Kc))
A025high3 <- vc - 0.5 * vo025high3 - R
vo05high3 <- Vomax * O2 / (O2 + Ko * (1 + Cc05high3 / Kc))
A05high3 <- vc - 0.5 * vo05high3 - R
vo1high3 <- Vomax * O2 / (O2 + Ko * (1 + Cc1high3 / Kc))
A1high3 <- vc - 0.5 * vo1high3 - R
vo2high3 <- Vomax * O2 / (O2 + Ko * (1 + Cc2high3 / Kc))
A2high3 <- vc - 0.5 * vo2high3 - R
vo4high3 <- Vomax * O2 / (O2 + Ko * (1 + Cc4high3 / Kc))
A4high3 <- vc - 0.5 * vo4high3 - R
Ahigh3 <-
c(A0125high3, A025high3, A05high3, A1high3, A2high3, A4high3)
Cidiffusion <- c(Cim0125, Cim025, Cim05, Cim1, Cim2, Cim4)
Alow <- c(A0125low, A025low, A05low, A1low, A2low, A4low)
Amedian <-
c(A0125median,
A025median,
A05median,
A1median,
A2median,
A4median)
Ahigh <- c(A0125high, A025high, A05high, A1high, A2high, A4high)
variable <- c(rep("Mesophyll Conductance", 6 * length(Cim1)))
conductance <-
c(rep(0.125, 376),
rep(0.25, 376),
rep(0.5, 376),
rep(1, 376),
rep(2, 376),
rep(4, 376))
Diffusionplot2 <-
as.data.frame(
cbind(
Cidiffusion,
Alow,
Amedian,
Ahigh,
Ahigh2,
Ahigh3,
variable,
conductance,
resistance
)
)
write.csv(Diffusionplot2, "./data/DiffusionLimitsACI2.csv")最终够到的不同导度下的扩散数据如下:
knitr::kable(head(Diffusionplot2))| Cidiffusion | Alow | Amedian | Ahigh | Ahigh2 | Ahigh3 | variable | conductance | resistance |
|---|---|---|---|---|---|---|---|---|
| -14.0805894452511 | -4.87461753940542 | -4.86419435199041 | -4.81536251060599 | -4.74710314038274 | -4.68025068475633 | Mesophyll Conductance | 0.125 | 11 |
| -11.8541114570766 | -4.72133615031493 | -4.71094118846005 | -4.66224099010836 | -4.59416400241068 | -4.52748832337504 | Mesophyll Conductance | 0.125 | 11 |
| -9.63096032171488 | -4.56847050260645 | -4.55810365181624 | -4.50953456487788 | -4.44163922971142 | -4.37513962701612 | Mesophyll Conductance | 0.125 | 11 |
| -7.41112252124025 | -4.41601890716257 | -4.40568005356001 | -4.35724154927421 | -4.28952714053524 | -4.22320291762348 | Mesophyll Conductance | 0.125 | 11 |
| -5.19458461086401 | -4.26397968400382 | -4.25366871432613 | -4.20536026677158 | -4.13782606222113 | -4.07167652620537 | Mesophyll Conductance | 0.125 | 11 |
| -2.98133321844053 | -4.11235116222692 | -4.10206796382144 | -4.05388904989726 | -3.98653433113539 | -3.9205587927733 | Mesophyll Conductance | 0.125 | 11 |
10.9 扩散限制作图
data <- read.csv("./data/DiffusionLimitsACI2.csv")
data$resistance <- as.factor(data$resistance)
graph <- ggplot(data, aes(x = Cidiffusion, y = Ahigh3, colour = resistance)) +
geom_abline(slope=0,intercept=0,size=1.5)+
geom_point()+
labs(colour = 'Total Resistance')+
scale_colour_brewer(palette = 'Spectral') +
theme_bw() +
scale_x_continuous(limits=c(0,100))+
scale_y_continuous(limits=c(-5,10))+
theme(
axis.title = element_text(size = 18),
axis.text = element_text(size = 15),
legend.position = 'right',
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()
)
graph## Warning: Removed 1863 rows containing missing values (geom_point).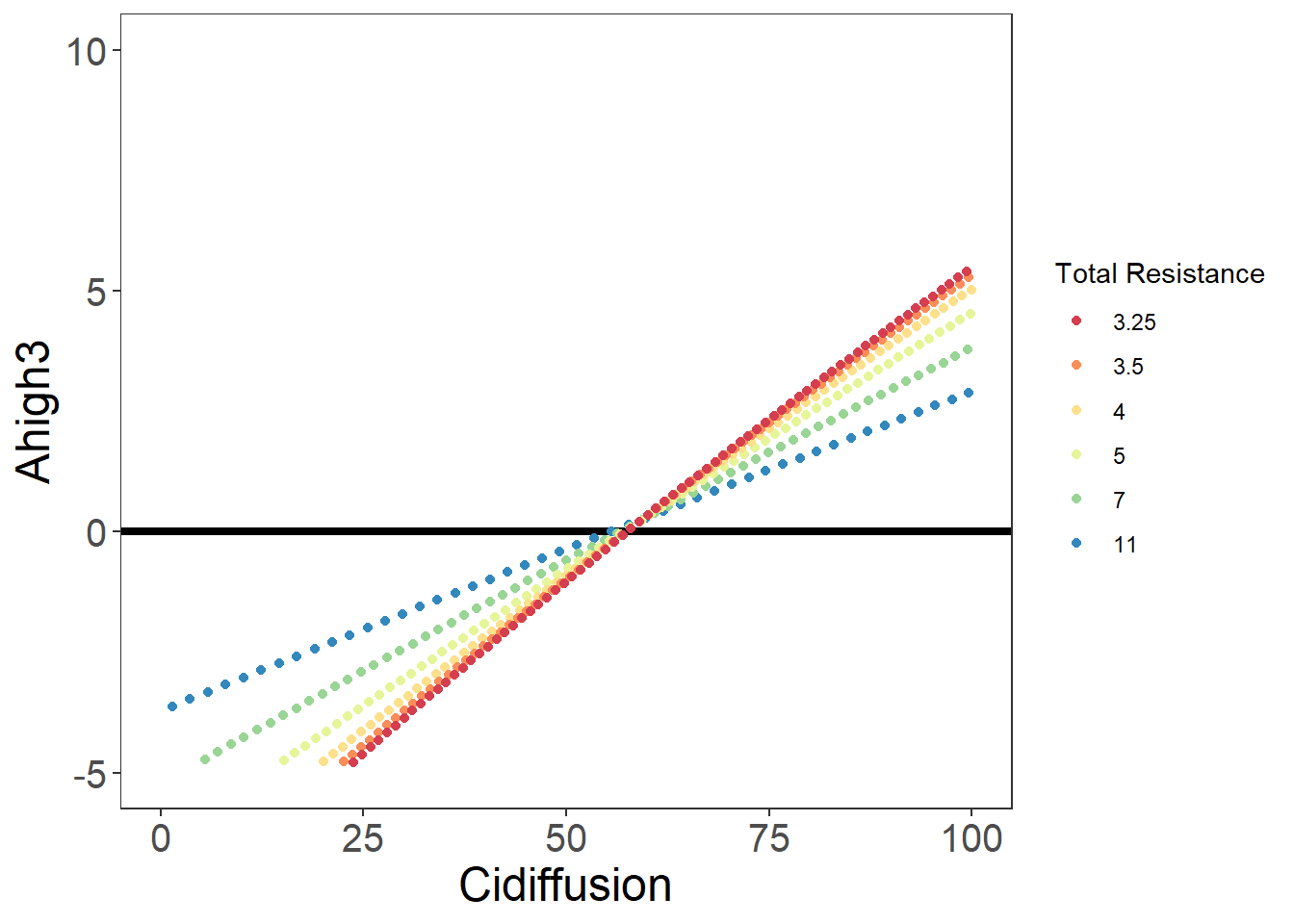
图 10.13: 不同 Ci 扩散限制下的差异
datahigh <- data[data$resistance == "11",]
graph <- ggplot(datahigh, aes(x = Cidiffusion, y = Ahigh3)) +
geom_abline(slope=0,intercept=0,size=1.5)+
geom_point(colour = "red")+
geom_point(aes(x = Cidiffusion, y = Ahigh2), colour = "blue")+
geom_point(aes(x = Cidiffusion, y = Ahigh), colour = "green")+
labs(colour = 'Total Resistance')+
scale_colour_brewer(palette = 'Spectral') +
theme_bw() +
scale_x_continuous(limits=c(0,100))+
scale_y_continuous(limits=c(-2.5,2.5))+
theme(
axis.title = element_text(size = 18),
axis.text = element_text(size = 15),
legend.position = 'right',
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()
)
graph## Warning: Removed 340 rows containing missing values (geom_point).
## Warning: Removed 340 rows containing missing values (geom_point).
## Warning: Removed 340 rows containing missing values (geom_point).
图 10.14: 不同 Ci 扩散限制下的差异(resistance = 11)
data2 <- read.csv("./data/DiffusionLimits.csv")
graph <- ggplot(data2, aes(x = TotalRes, y = Rates, colour = Scenario)) +
geom_point()+
scale_colour_brewer(palette = 'Spectral') +
theme_bw() +
theme(
axis.title = element_text(size = 18),
axis.text = element_text(size = 12),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()
)
graph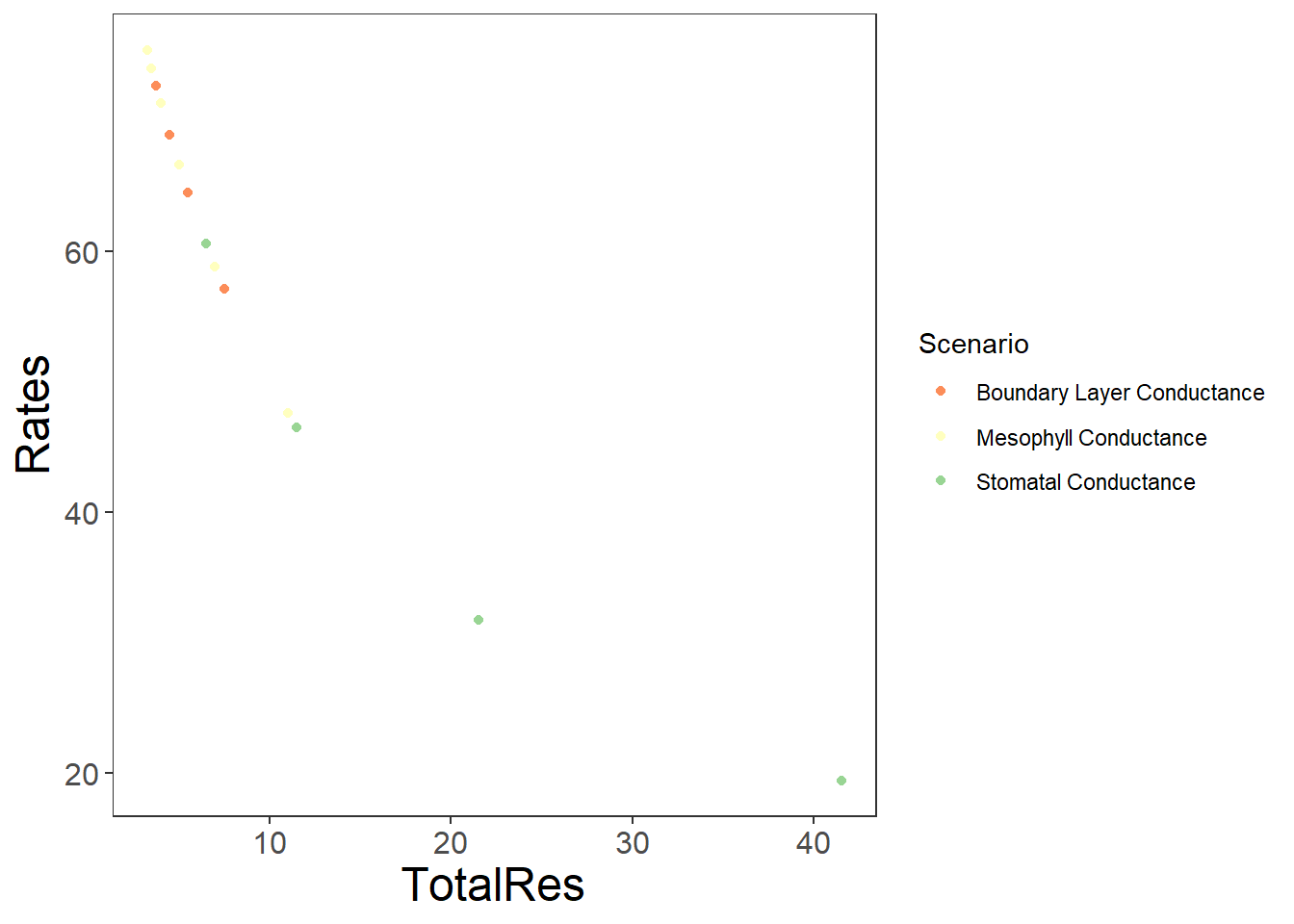
图 10.15: 不同总导度下的各个导度的速率变化
m1 <- lm(Rates ~ I(1/TotalRes)+I((1/TotalRes)^2),data=data2)
summary(m1)##
## Call:
## lm(formula = Rates ~ I(1/TotalRes) + I((1/TotalRes)^2), data = data2)
##
## Residuals:
## Min 1Q Median 3Q Max
## -3.3590 -0.9844 -0.1236 0.7706 2.4510
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 13.042 1.684 7.745 3.18e-06 ***
## I(1/TotalRes) 421.928 22.487 18.763 8.49e-11 ***
## I((1/TotalRes)^2) -737.690 65.500 -11.262 4.46e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.705 on 13 degrees of freedom
## Multiple R-squared: 0.9904, Adjusted R-squared: 0.9889
## F-statistic: 668.8 on 2 and 13 DF, p-value: 7.799e-14ab <- predict(m1)
red <- 1/data2$TotalRes
plot(ab~red)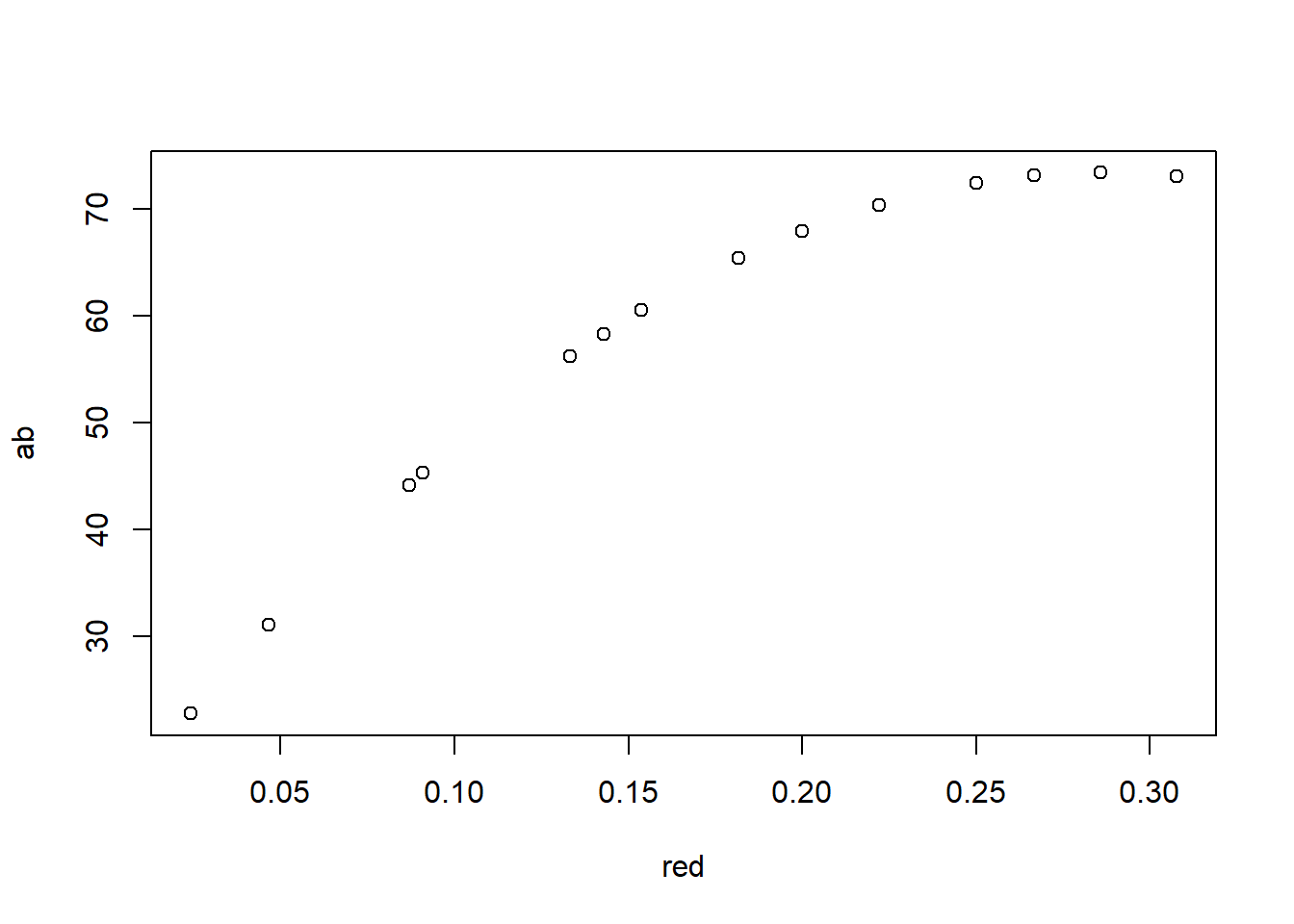
图 10.16: 不同阻力下的各个导度的速率变化预测值
10.9.1 补偿点的计算
计算补偿点,代码同前文类似,只是采用了不同导度下的数值:
data <- read.csv("./data/DiffusionLimitsACI2.csv")
#Gamma
# For Ci-based estimates, only use Ci < 100
dataCi <- data[data$Cidiffusion < 100, ]
dataCi0125 <- dataCi[dataCi$conductance == "0.125", ]
dataCi025 <- dataCi[dataCi$conductance == "0.25", ]
dataCi05 <- dataCi[dataCi$conductance == "0.5", ]
dataCi1 <- dataCi[dataCi$conductance == "1", ]
dataCi2 <- dataCi[dataCi$conductance == "2", ]
dataCi4 <- dataCi[dataCi$conductance == "4", ]
m1 <- lm(dataCi0125$Ahigh ~ dataCi0125$Cidiffusion)
summary(m1)##
## Call:
## lm(formula = dataCi0125$Ahigh ~ dataCi0125$Cidiffusion)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.040649 -0.014495 0.005307 0.017054 0.021232
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -3.836e+00 4.318e-03 -888.5 <2e-16 ***
## dataCi0125$Cidiffusion 6.665e-02 7.873e-05 846.6 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.01934 on 52 degrees of freedom
## Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
## F-statistic: 7.167e+05 on 1 and 52 DF, p-value: < 2.2e-16Gamma0125 <- -m1$coefficients[1] / m1$coefficients[2]
m2 <- lm(dataCi025$Ahigh ~ dataCi025$Cidiffusion)
summary(m2)##
## Call:
## lm(formula = dataCi025$Ahigh ~ dataCi025$Cidiffusion)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.070876 -0.025427 0.009071 0.029765 0.036593
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -5.2507346 0.0092187 -569.6 <2e-16 ***
## dataCi025$Cidiffusion 0.0904227 0.0001544 585.5 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.03329 on 59 degrees of freedom
## Multiple R-squared: 0.9998, Adjusted R-squared: 0.9998
## F-statistic: 3.428e+05 on 1 and 59 DF, p-value: < 2.2e-16Gamma025 <- -m2$coefficients[1] / m2$coefficients[2]
m3 <- lm(dataCi05$Ahigh ~ dataCi05$Cidiffusion)
summary(m3)##
## Call:
## lm(formula = dataCi05$Ahigh ~ dataCi05$Cidiffusion)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.10435 -0.03583 0.01358 0.04311 0.05340
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -6.4051036 0.0150363 -426 <2e-16 ***
## dataCi05$Cidiffusion 0.1097686 0.0002391 459 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.04851 on 65 degrees of freedom
## Multiple R-squared: 0.9997, Adjusted R-squared: 0.9997
## F-statistic: 2.107e+05 on 1 and 65 DF, p-value: < 2.2e-16Gamma05 <- -m3$coefficients[1] / m3$coefficients[2]
m4 <- lm(dataCi1$Ahigh ~ dataCi1$Cidiffusion)
summary(m4)##
## Call:
## lm(formula = dataCi1$Ahigh ~ dataCi1$Cidiffusion)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.13150 -0.04513 0.01709 0.05402 0.06692
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -7.1817472 0.0199135 -360.6 <2e-16 ***
## dataCi1$Cidiffusion 0.1227771 0.0003085 398.0 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.06074 on 69 degrees of freedom
## Multiple R-squared: 0.9996, Adjusted R-squared: 0.9996
## F-statistic: 1.584e+05 on 1 and 69 DF, p-value: < 2.2e-16Gamma1 <- -m4$coefficients[1] / m4$coefficients[2]
m5 <- lm(dataCi2$Ahigh ~ dataCi2$Cidiffusion)
summary(m5)##
## Call:
## lm(formula = dataCi2$Ahigh ~ dataCi2$Cidiffusion)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.14803 -0.04874 0.01837 0.05995 0.07511
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -7.6427727 0.0230595 -331.4 <2e-16 ***
## dataCi2$Cidiffusion 0.1305083 0.0003538 368.9 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.06815 on 71 degrees of freedom
## Multiple R-squared: 0.9995, Adjusted R-squared: 0.9995
## F-statistic: 1.361e+05 on 1 and 71 DF, p-value: < 2.2e-16Gamma2 <- -m5$coefficients[1] / m5$coefficients[2]
m6 <- lm(dataCi4$Ahigh ~ dataCi4$Cidiffusion)
summary(m6)##
## Call:
## lm(formula = dataCi4$Ahigh ~ dataCi4$Cidiffusion)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.15721 -0.05258 0.01994 0.06470 0.07968
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -7.8955772 0.0248634 -317.6 <2e-16 ***
## dataCi4$Cidiffusion 0.1347503 0.0003799 354.7 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.07225 on 72 degrees of freedom
## Multiple R-squared: 0.9994, Adjusted R-squared: 0.9994
## F-statistic: 1.258e+05 on 1 and 72 DF, p-value: < 2.2e-16Gamma4 <- -m6$coefficients[1] / m6$coefficients[2]
GammaCi <- c(Gamma0125, Gamma025, Gamma05, Gamma1, Gamma2, Gamma4)10.9.2 所有图形代码
data <- read.csv("./data/PRdata.csv")
data$Ccfull <- as.numeric(data$Ccfull)
data$Delay <- as.factor(data$Delay)
cols <- gg_color_hue(6)
Panel_1A <-
ggplot(data, aes(
x = Cifull,
y = Anet,
colour = Delay,
linetype = Delay
)) +
# 零水平参考线
geom_abline(slope = 0,
intercept = 0,
size = 1.5) +
#ci与A loess 方法的拟合曲线
geom_smooth(method = "loess", se = FALSE, size = 2) +
scale_linetype_manual(
name = "Delay (s)",
labels = c("0", "15", "30", "60", "120", "300"),
values = c("solid", "longdash", "twodash", "dotdash", "dashed", "dotted")
) +
ggtitle(expression(paste(
bold("(a)"), " Modelled Photorespiratory Effect"
))) +
labs(x = expression(C[i] ~ "(" * mu * mol ~ mol ^ {
-1
} * ")"),
y = expression(A[net] ~ "(" * mu * mol ~ m ^ {
-2
} ~ s ^ {
-1
} * ")")) +
labs(colour = 'Delay (s)') +
scale_x_continuous(limits = c(0, 250)) +
scale_y_continuous(limits = c(-5, 20)) +
scale_colour_manual(
values = cols,
name = "Delay (s)",
labels = c("0", "15", "30", "60", "120", "300")
) +
theme_bw() +
theme(
axis.title.x = element_text(size = 20),
axis.title.y = element_text(size = 20),
axis.line = element_line(size = 1),
axis.ticks = element_line(size = 1.5),
axis.ticks.length = unit(1.5, "mm"),
rect = element_rect(size = 2),
axis.text.x = element_text(size = 14, color = 'black'),
axis.text.y =
element_text(
size = 14,
color = 'black',
hjust = (1)
),
legend.position = c(0.15, 0.75),
axis.title = element_text(size = 18),
axis.text = element_text(size = 12, color = "black"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.text = element_text(size = 14),
legend.title = element_text(size = 14)
)
Panel_1A## Warning: Removed 1029 rows containing non-finite values (stat_smooth).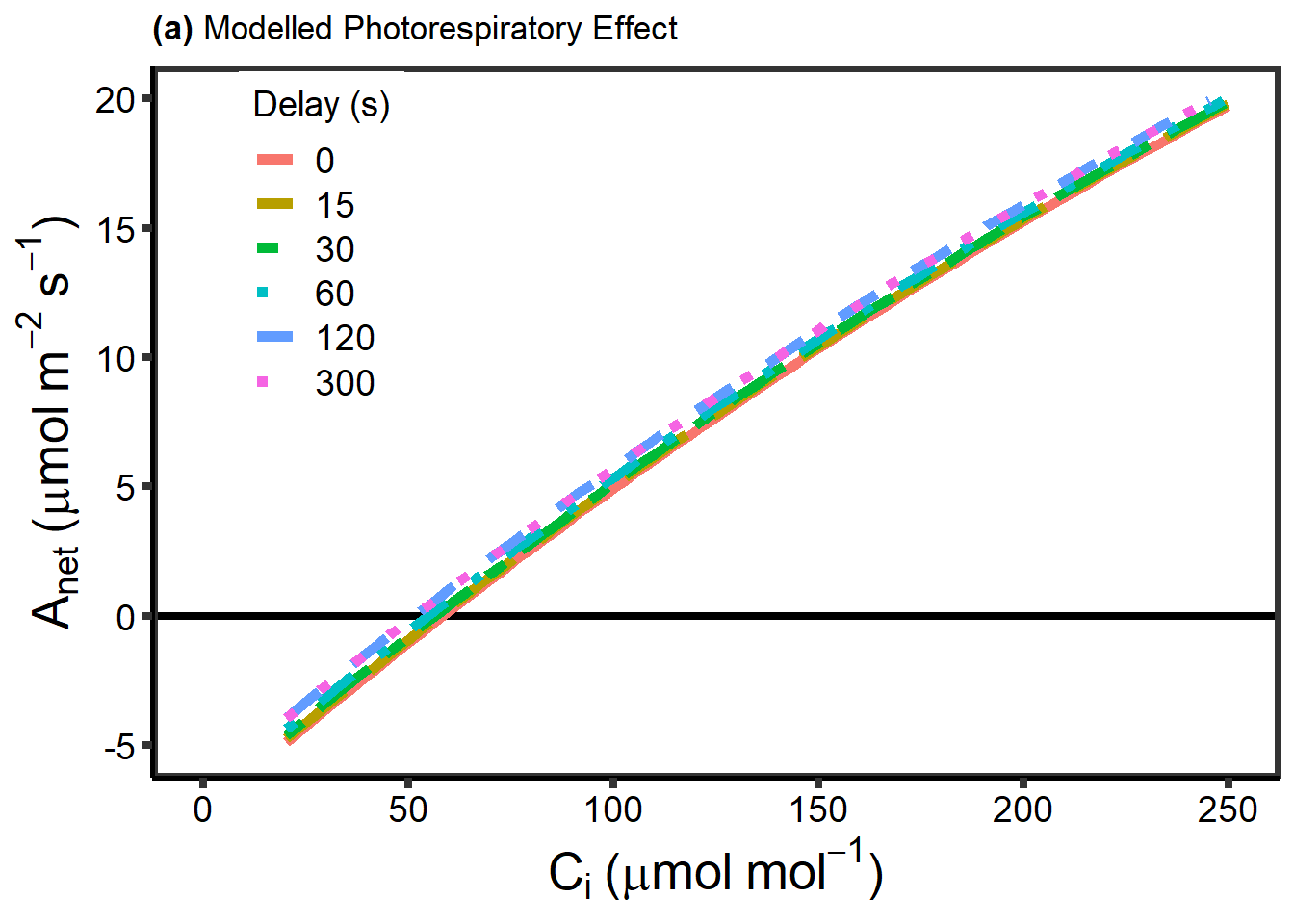
图 10.17: 不同时间滞后性的 Anet VS Ci
Panel_1A_inset <-
ggplot(data, aes(
x = Cifull,
y = Anet,
colour = Delay,
linetype = Delay
)) +
geom_abline(slope = 0,
intercept = 0,
size = 1.5) +
geom_smooth(method = "loess", se = FALSE, size = 2) +
scale_linetype_manual(
name = "Delay (s)",
labels = c("0", "15", "30", "60", "120", "300"),
values = c("solid", "longdash", "twodash", "dotdash", "dashed", "dotted")
) +
labs(x = expression(C[i] ~ "(" * mu * mol ~ mol ^ {
-1
} * ")"),
y = expression(A[net] ~ "(" * mu * mol ~ m ^ {
-2
} ~ s ^ {
-1
} * ")")) +
labs(colour = 'Delay (s)') +
scale_x_continuous(limits = c(45, 65)) +
scale_y_continuous(limits = c(-1, 1), breaks = c(-1, 0, 1)) +
scale_colour_manual(values = cols) +
theme_bw() +
theme(
axis.title.x = element_blank(),
axis.title.y = element_blank(),
axis.line = element_line(size = 1),
axis.ticks = element_line(size = 1.5),
axis.ticks.length = unit(1.5, "mm"),
rect = element_rect(size = 2),
axis.text.x = element_text(size = 14, color = 'black'),
axis.text.y =
element_text(
size = 14,
color = 'black',
hjust = (1)
),
axis.title = element_blank(),
axis.text = element_text(size = 12, color = "black"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = 'none',
plot.background = element_blank()
)
cols <- gg_color_hue(6)
data <- read.csv("./data/DiffusionLimitsACI2.csv")
data$resistance <- as.factor(data$resistance)
Panel_1Db <-
ggplot(data,
aes(
x = Cidiffusion,
y = Ahigh,
colour = resistance,
linetype = resistance
)) +
geom_abline(slope = 0,
intercept = 0,
size = 1.5) +
geom_smooth(method = "loess", se = FALSE, size = 2) +
scale_linetype_manual(
name = "Total Resistance",
labels = c("3.25", "3.5", "4", "5", "7", "11"),
values = c("solid", "longdash", "twodash", "dotdash", "dashed", "dotted")
) +
ggtitle(expression(paste(bold("(d)"), " Modelled Resistance Effect"))) +
scale_color_manual(
values = cols,
name = "Total Resistance",
labels = c("3.25", "3.5", "4", "5", "7", "11")
) +
theme_bw() +
labs(x = expression(paste(C[i] ~ "(" * mu * mol ~ mol ^ {
-1
} * ")")), y = expression(paste(A[net] ~ "(" * mu * mol ~ m ^ {
-2
} ~ s ^ {
-1
} * ")"))) +
scale_x_continuous(limits = c(0, 250)) +
scale_y_continuous(limits = c(-5, 20)) +
theme(
axis.title.x = element_text(size = 20),
axis.title.y = element_text(size = 20),
axis.line = element_line(size = 1),
axis.ticks = element_line(size = 1.5),
axis.ticks.length = unit(1.5, "mm"),
rect = element_rect(size = 2),
axis.text.x = element_text(size = 14, color = 'black'),
axis.text.y =
element_text(
size = 14,
color = 'black',
hjust = (1)
),
legend.position = c(0.22, 0.75),
axis.title = element_text(size = 18),
axis.text = element_text(size = 12, color = "black"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.text = element_text(size = 14),
legend.title = element_text(size = 14)
)
Panel_1Db## Warning: Removed 1159 rows containing non-finite values (stat_smooth).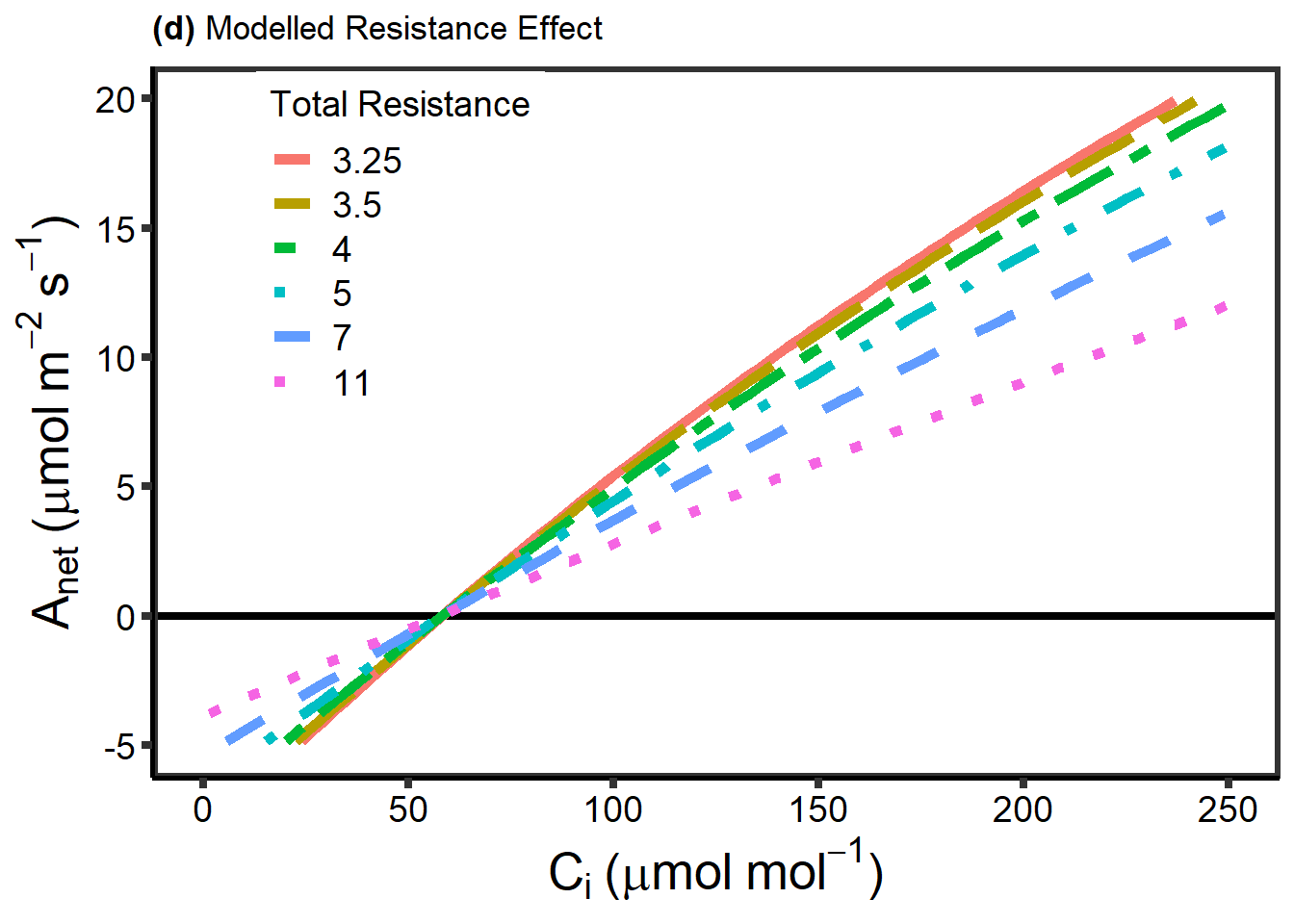
图 10.18: 不同总阻力下的 Anet VS Ci
data <- read.csv("./data/DiffusionLimitsACI2.csv")
data$resistance <- as.factor(data$resistance)
data <- data[data$resistance == "11",]
resistance <- rep(data$resistance, 3)
Cidiffusion <- rep(data$Cidffusion, 3)
A <- c(data$Ahigh, data$Ahigh2, data$Ahigh3)
Rate <-
c(rep(100, length(data$Ahigh)), rep(200, length(data$Ahigh2)), rep(300, length(data$Ahigh3)))
data3 <- rbind(data, data, data)
data3$A <- c(data$Ahigh, data$Ahigh2, data$Ahigh3)
data3$Rate <-
as.factor(c(rep(100, length(data$Ahigh)), rep(200, length(data$Ahigh2)), rep(300, length(data$Ahigh3))))
Panel_1Dbinset <-
ggplot(data3, aes(
x = Cidiffusion,
y = A,
colour = Rate,
linetype = Rate
)) +
geom_abline(slope = 0,
intercept = 0,
size = 1.5) +
geom_smooth(method = "loess", se = FALSE, size = 2) +
scale_linetype_manual(
name = "Rate",
labels = c("100", "200", "300"),
values = c("solid", "longdash", "twodash")
) +
scale_color_manual(
values = cols,
name = "Rate",
labels = c("100", "200", "300")
) +
theme_bw() +
labs(x = expression(paste(C[i] ~ "(" * mu * mol ~ mol ^ {
-1
} * ")")), y = expression(paste(A[net] ~ "(" * mu * mol ~ m ^ {
-2
} ~ s ^ {
-1
} * ")"))) +
scale_x_continuous(limits = c(45, 65)) +
scale_y_continuous(limits = c(-1, 1), breaks = c(-1, 0, 1)) +
theme(
axis.title.x = element_blank(),
axis.title.y = element_blank(),
axis.line = element_line(size = 1),
axis.ticks = element_line(size = 1.5),
axis.ticks.length = unit(1.5, "mm"),
rect = element_rect(size = 2),
axis.text.x = element_text(size = 14, color = 'black'),
axis.text.y =
element_text(
size = 14,
color = 'black',
hjust = (1)
),
legend.position = c(0.22, 0.75),
axis.title = element_text(size = 18),
axis.text = element_text(size = 12, color = "black"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
plot.background = element_blank(),
legend.text = element_text(size = 12),
legend.key = element_blank(),
legend.title = element_blank(),
legend.background = element_blank()
)
Panel_1Dbinset## Warning: Removed 1101 rows containing non-finite values (stat_smooth).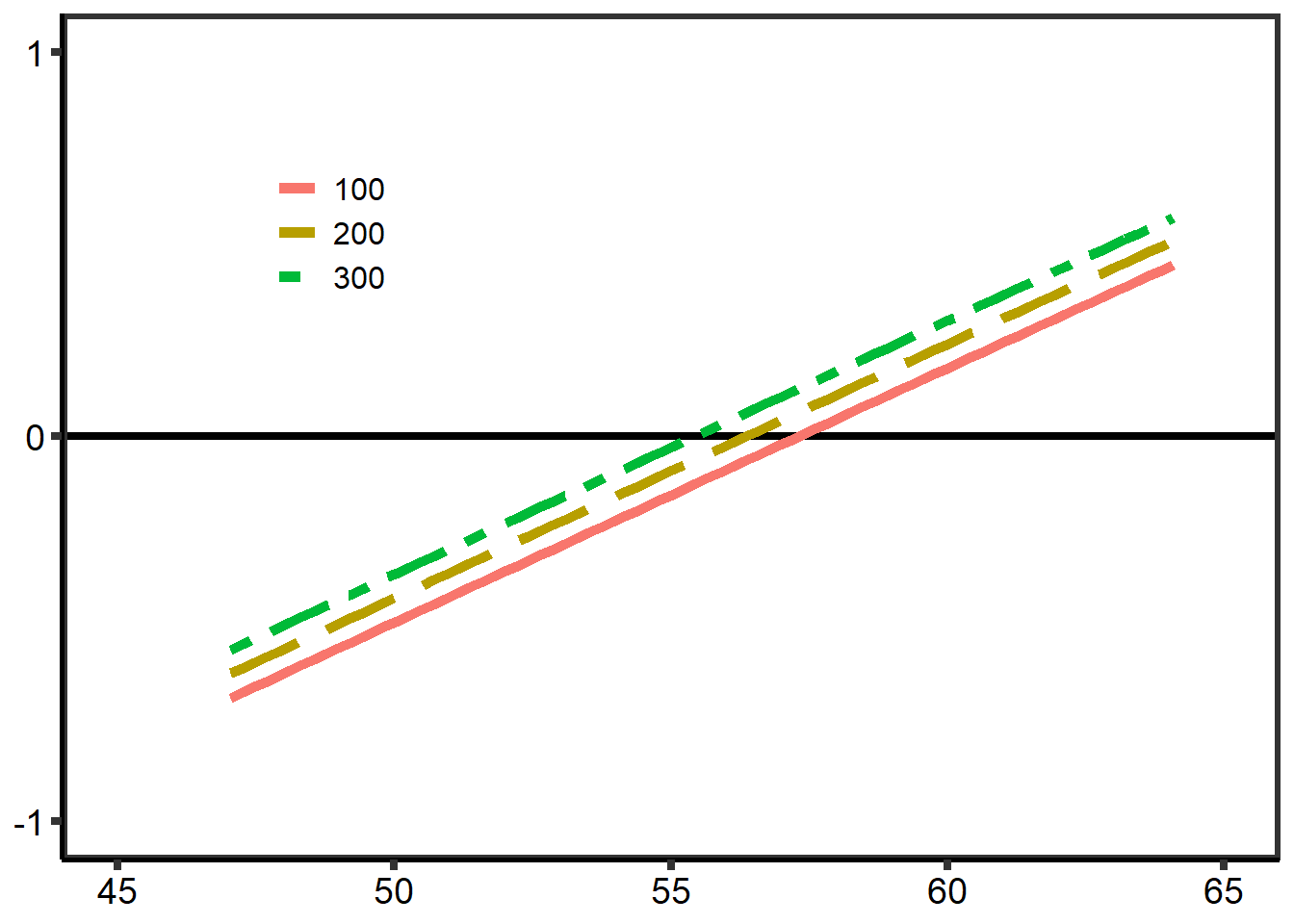
图 10.19: 不同导度下的 A VS Ci
data2 <- read.csv("./data/DiffusionLimits.csv")
data2$Conductance <-
revalue(
data2$Scenario,
c(
"Boundary Layer Conductance" = "Boundary Layer",
"Mesophyll Conductance" = "Mesophyll",
"Stomatal Conductance" = "Stomatal"
)
)
cols <- gg_color_hue(3)
Panel_1E <-
ggplot(data2, aes(x = TotalRes, y = Rates, colour = Conductance)) +
geom_point(size = 4) +
ggtitle(expression(paste(bold("(e)"), " Modelled Resistance Effect"))) +
labs(x = expression(paste(r[total] * " (s" ~ m ^ {
2
} ~ mol ^ {
-1
} * ")")),
y = expression(paste(C[c] * " Ramp Rate (" * mu * mol ~ mol ^
{
-1
} ~ min ^ {
-1
} * ")"))) +
scale_x_continuous(limits = c(0, 50)) +
scale_y_continuous(limits = c(0, 80)) +
scale_color_manual(
values = cols,
name = "Manipulated Resistance",
labels = c("Boundary Layer", "Mesophyll", "Stomatal")
) +
theme_bw() +
theme(
axis.title.x = element_text(size = 20),
axis.title.y = element_text(size = 20),
axis.line = element_line(size = 1),
axis.ticks = element_line(size = 1.5),
axis.ticks.length = unit(1.5, "mm"),
rect = element_rect(size = 2),
axis.text.x = element_text(size = 14, color = 'black'),
axis.text.y =
element_text(
size = 14,
color = 'black',
hjust = (1)
),
legend.position = c(0.70, 0.84),
axis.title = element_text(size = 18),
axis.text = element_text(size = 12, color = "black"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.text = element_text(size = 14),
legend.title = element_text(size = 14)
)
Panel_1E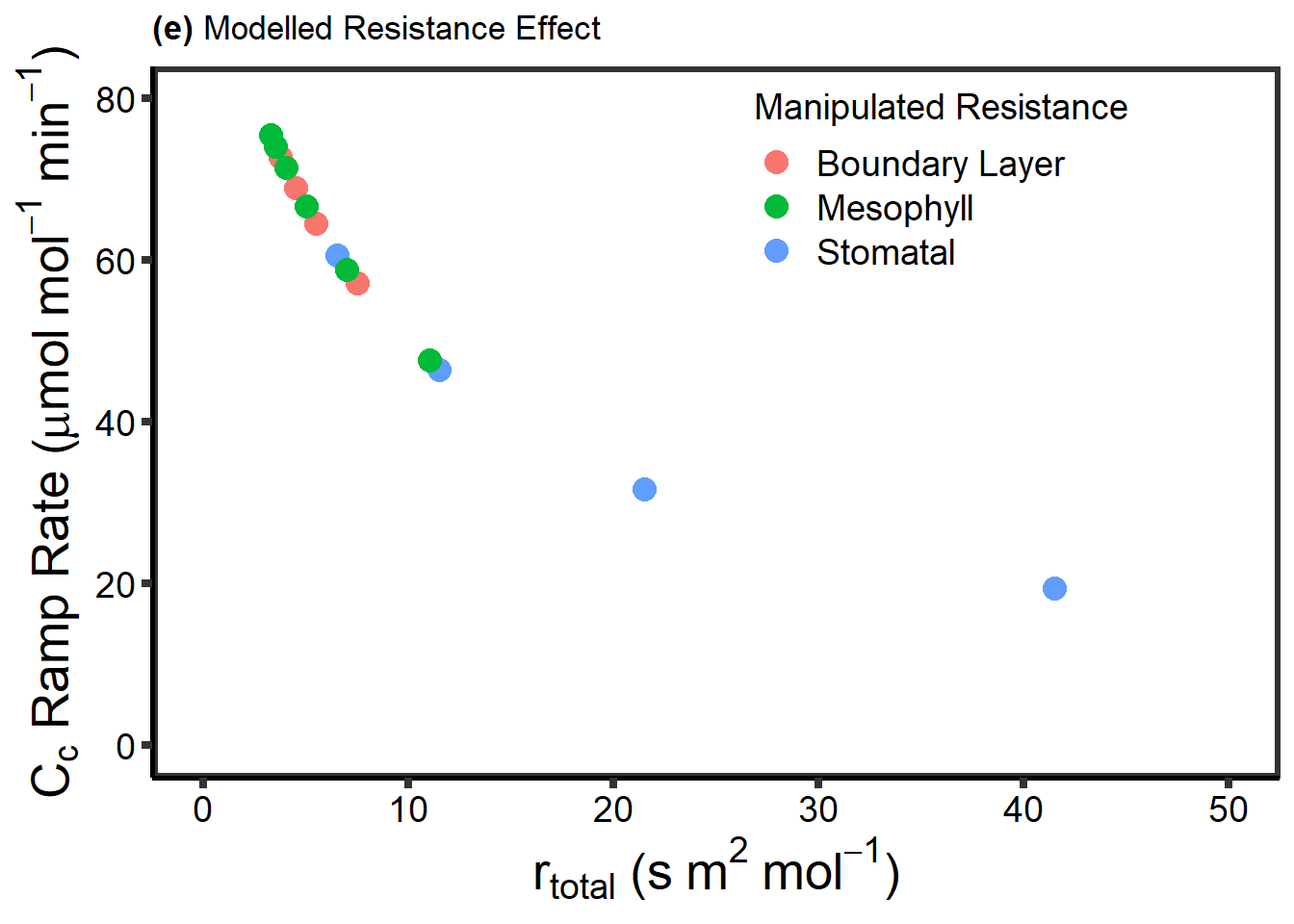
图 10.20: 不同总导度下的各个导度的变化
data <- read.csv("./data/RASdata.csv")
data$Ccfull <- as.numeric(data$Ccfull)
data$Deactivation <- as.factor(data$Deactivation)
print(levels(data$Deactivation))## [1] "High" "Low" "Medium" "None"data$Deactivation <-
factor(data$Deactivation, levels(data$Deactivation)[c(4, 2, 3, 1)])
FigS1 <-
ggplot(data,
aes(
x = Cifull,
y = Anet,
colour = Deactivation,
linetype = Deactivation
)) +
geom_smooth(method = "loess", se = FALSE, size = 2) +
scale_linetype_manual(
name = "Deactivation",
labels = c("None", "Low", "Medium", "High"),
values = c("solid", "longdash", "dashed", "dotted")
) +
labs(x = expression(C[i] ~ "(" * mu * mol ~ mol ^ {
-1
} * ")"),
y = expression(A[net] ~ "(" * mu * mol ~ m ^ {
-2
} ~ s ^ {
-1
} * ")")) +
labs(colour = 'Deactivation') +
scale_x_continuous(limits = c(25, 100),
breaks = c(25, 40, 55, 70, 85, 100)) +
scale_y_continuous(limits = c(-5, 5)) +
geom_hline(yintercept = 0, size = 1.5) +
theme_bw() +
theme(
axis.title.x = element_text(size = 20),
axis.title.y = element_text(size = 20),
axis.line = element_line(size = 1),
axis.ticks = element_line(size = 1.5),
axis.ticks.length = unit(1.5, "mm"),
rect = element_rect(size = 2),
axis.text.x = element_text(size = 14, color = 'black'),
axis.text.y =
element_text(
size = 14,
color = 'black',
hjust = (1)
),
legend.position = c(0.8, 0.2),
legend.text = element_text(size = 16),
legend.title = element_text(size = 16),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()
)
FigS1## Warning: Removed 1230 rows containing non-finite values (stat_smooth).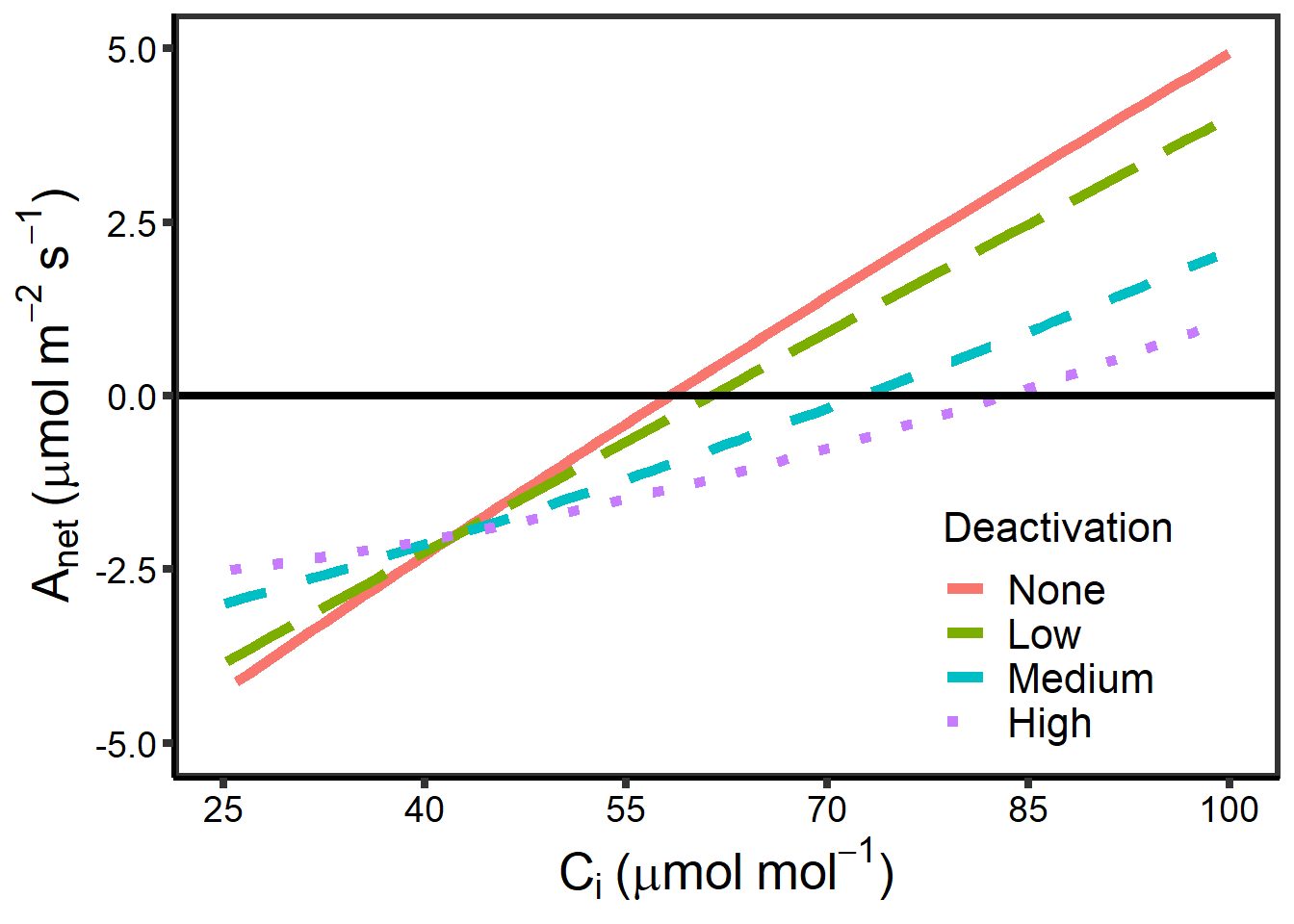
图 10.21: Rubisco不同失活程度下的下 A VS Ci
10.10 LI-6800 荧光数据分析
LI-6800 能够直接提供基本的叶绿素荧光参数,其他参数均可通过这些基本的参数进行计算,计算也较简单,在此不赘述,需要注意的是快相荧光部分的数据,因为分析 ojip 数据的模型有很多,很多都需要复杂的计算,在此我们先将其中较为简单的 jip test 数据分析进行介绍。
10.10.1 jip test 的实现
LI-6800 增加了 ojip 曲线测量功能,本功能主要是针对测量数据的 jip test 的实现。
10.10.2 jiptest 软件包安装
请在 github 安装,有可能需要安装 remotes
install.packages("remotes")然后安装:
install_github("zhujiedong/jiptest")如果因为网络原因失败,可以转战 gitee:
install_git('https://gitee.com/zhu_jie_dong/jiptest')10.10.3 read_files 及 read_dcfiles 函数
read_files 用于批量读取所有调制光测量数据,方便用于其他的数据分析。函数要求所有数据必须是 xlsx 格式,并且所有测量数据都保存在同一文件夹内。,如有其他文件或测量数据则会报错。
read_dcfiles 用于批量读取所有连续光测量数据,其他与 read_dcfiles 相同。例如我放在了 data文件夹下的 ojip 文件夹内,有五个数据文件。
函数仅有一个参数,即保存数据文件夹的路径,使用如下:
library(jiptest)
jip_data<- read_files("data/ojip")jip_dcdata <- read_dcfiles("./data/ojip")调制光的信号前 10 行数据:
| SECS | FLUOR | NORM_FLUOR | SOURCE |
|---|---|---|---|
| 6.10e-06 | 621 | 0.0005027 | INDUCTION-4188-20201116-10_53_39 |
| 1.01e-05 | 620 | 0.0000000 | INDUCTION-4188-20201116-10_53_39 |
| 1.42e-05 | 649 | 0.0145781 | INDUCTION-4188-20201116-10_53_39 |
| 1.80e-05 | 629 | 0.0045242 | INDUCTION-4188-20201116-10_53_39 |
| 2.21e-05 | 674 | 0.0271455 | INDUCTION-4188-20201116-10_53_39 |
| 2.61e-05 | 736 | 0.0583126 | INDUCTION-4188-20201116-10_53_39 |
连续光的信号前 10 行数据:
| SECS | FLUOR | NORM_FLUOR | SOURCE |
|---|---|---|---|
| 6.10e-06 | 98161 | 0.0000000 | INDUCTION-4188-20201116-10_53_39 |
| 1.01e-05 | 100331 | 0.0071347 | INDUCTION-4188-20201116-10_53_39 |
| 1.42e-05 | 102577 | 0.0145193 | INDUCTION-4188-20201116-10_53_39 |
| 1.80e-05 | 104849 | 0.0219894 | INDUCTION-4188-20201116-10_53_39 |
| 2.21e-05 | 107214 | 0.0297652 | INDUCTION-4188-20201116-10_53_39 |
| 2.61e-05 | 109624 | 0.0376890 | INDUCTION-4188-20201116-10_53_39 |
注:NORM_FLUOR 是标准化后的荧光信号数据,其中标准化采用的方式是 Stirbet and Govindjee (2012) 所采用的:\(F = \frac{F_t - F_o}{F_m - F_o}\),而曲线上部的面积则采用 Stirbet and Govindjee (2011) 的方式进行标准化,以方便不同叶片之间的比较,具体为 \(area = \frac{area}{F_m - F_o}\)
10.10.4 jip_test 及 jip_dctest 函数
jiptest 是 jiptest 的核心函数,用于所有数据的 jiptest 分析,函数仅包一个参数,测量数据的保存文件夹路径。
jip_dctest 与 jip_test 相似,用于连续光测量数据的分析。
actest <- jip_test("./data/ojip")dctest <- jip_dctest("./data/ojip")可以看出,对于标准化之后的参数,二者基本一致,原始信号则无法一致,因为连续光信号强度更高(所以数据的比较要在相同的测量光内进行):
调制光信号的计算参数:
| parameters | data_file1 | data_file2 | data_file3 | data_file4 | data_file5 |
|---|---|---|---|---|---|
| Fo | 620.000 | 887.000 | 849.000 | 981.000 | 914.000 |
| Fm | 2609.280 | 4649.520 | 3714.270 | 4225.240 | 3754.270 |
| F300 | 1593.200 | 2181.400 | 1798.600 | 2067.800 | 2537.200 |
| FJ | 1722.900 | 2594.300 | 2104.600 | 2456.800 | 2569.000 |
| FI | 2268.950 | 4443.110 | 3547.110 | 4009.220 | 3556.530 |
| Tfmax | 159.998 | 256.016 | 316.016 | 272.016 | 200.016 |
连续光信号计算参数:
| parameters | data_file1 | data_file2 | data_file3 | data_file4 | data_file5 |
|---|---|---|---|---|---|
| Fo | 98161.000 | 154000.000 | 133191.000 | 156664.000 | 153589.000 |
| Fm | 402308.000 | 713839.000 | 560610.000 | 647660.000 | 584839.000 |
| F300 | 246753.000 | 338091.000 | 275476.800 | 322859.200 | 388110.400 |
| FJ | 267827.000 | 400744.000 | 320245.000 | 376885.000 | 406671.000 |
| FI | 349985.000 | 681505.000 | 534314.000 | 614590.000 | 552253.000 |
| Tfmax | 171.998 | 276.016 | 300.016 | 288.016 | 212.016 |
计算参数见表 10.3 及 10.4,考虑到排版,仅显示部分内容。若需要将数据导出,可以使用相关命令,如:
# export the results of jiptest to a csv file
write.csv(actest, "d:/data/jip_resluts.csv")
write.csv(dctest, "d:/data/dcjip_resluts.csv")10.10.5 图像查看函数
之前的预览方式已经彻底从代码中删除,不在提供。如果仅仅是简单查看结果,那么我们直接用默认参数就好,作图的数据是之前导入的数据文件:
默认使用标准化的荧光信号(也就是除以最大值,标准化到 0~1 之间,LI-6800 在 1.4 之后的版本也提供了仪器上查看的界面)。
10.10.5.1 默认图形的方式
- 默认调制光图形
plot(jip_data)
图 10.22: 默认调制光图形查看
- 默认连续光图形
plot(jip_dcdata)
图 10.23: 默认连续光图形查看
默认图形,仅仅查看结果是可以的,当然有些情况下需要进行调整才方便做展示用。
10.10.5.2 定制化图形的方式
这个是这次改动的重点之一,例如都使用原始信号分别做上面两幅图:
- 定制调制光图形
这里使用原始荧光信号,然后更改图例颜色等,主要是 add_leg = FALSE 不增加默认的图例,以及 normalized = FALSE 不使用信号标准化。其他请参考帮助 ?plot.jip。 我简单写了一下帮助文档,大家应该基本能看明白,我实在没时间。
cls <- palette.colors(n = 5, "set 2", alpha = 0.8)
plot(jip_data,
ylab = 'Normalized fluorescence signals',
add_leg = FALSE,
def_pch = 14:18,
col = cls,
main = "Demodulated signals", normalized = FALSE)
legend(
"topleft",
unique(jip_data$SOURCE),
col = cls,
pch = 14:18,
cex = 0.6,
pt.cex = 1.2,
bty = "n")
图 10.24: 定制调制光图形示例
- 定制连续光图形
连续光图形,只改了两个地方,图例的位置以及不显示标准化的数据。
plot(jip_dcdata, legend_pos = "bottomright", normalized = FALSE)
图 10.25: 定制连续光图形示例
关于标准化和非标准化图形差异大的问题,这个比较简单,原始信号比较低的那条线,是不同物种的不同叶片,所以不具备比较价值,我只是数据测试。
建议在开始分析数据前,使用如上方式作图查看数据质量,若使用调制光数据测量的荧光信号太弱,数据点太散,则可以使用连续光测量信号进行分析,对于归一化的荧光参数,二者几乎无差别,当然避免陷入被动的最好方式还是最开始测量时注意检查数据质量